More details and advanced usage of the `vaeac` approach
Lars Henry Berge Olsen
Source:vignettes/vaeac.Rmd
vaeac.RmdIn this vignette, we elaborate and illustrate the vaeac
approach in more depth than in the general usage vignette. In the
general usage vignette, only a few basic examples of using
vaeac are included, while we showcase more advanced usage
here. See the overview above for which topics are covered in this
vignette.
Vaeac
An approach that supports mixed features is the Variational
AutoEncoder with Arbitrary Conditioning (Olsen et
al. (2022)), abbreviated to vaeac. The
vaeac is an extension of the regular variational
autoencoder (Kingma and Welling (2014)),
but instead of giving a probabilistic representation of the distribution
it gives a probabilistic representation of the conditional distribution
,
for all possible feature subsets
simultaneously, where
is the set of all features. That is, only a single vaeac
model is needed to model all conditional distributions.
The vaeac consists of three neural networks: a full
encoder, a masked encoder, and a decoder. The
encoders map the full and masked/conditional input representations,
i.e.,
and
,
respectively, to latent probabilistic representations. Sampled instances
from these latent probabilistic representations are sent to the decoder,
which maps them back to the feature space and provides a sampleable
probabilistic representation for the unconditioned features
.
The full encoder is only used during the training phase of the
vaeac model to guide the training process of the masked
encoder, as the former relies on the full input sample
,
which is not accessible in the deployment phase (when we generate the
Monte Carlo samples), as we only have access to
.
The networks are trained by minimizing a variational lower bound, and
see Section 3 in Olsen et al. (2022) for
an in-depth introduction to the vaeac methodology. We use
the vaeac model at the epoch which obtains the lowest
validation IWAE score to generate the Monte Carlo samples used in the
Shapley value computations.
We fit the vaeac model using the torch package
in
(Falbel and Luraschi (2023)). The main
parameters are the number of layers in the networks
(vaeac.depth), the width of the layers
(vaeac.width), the number of dimensions in the latent space
(vaeac.latent_dim), the activation function between the
layers in the networks (vaeac.activation_function), the
learning rate in the ADAM optimizer (vaeac.lr), the number
of vaeac models to initiate to remedy poorly initiated
model parameter values (vaeac.n_vaeacs_initialize), and the
number of learning epochs (vaeac.epochs). Call
?shapr::setup_approach.vaeac for a more detailed
description of the parameters.
There are additional extra parameters which can be set by including a
named list in the call to the explain() function. For
example, we can change the batch size to 32 by including
vaeac.extra_parameters = list(vaeac.batch_size = 32) as a
parameter in the call to the explain() function. See
?shapr::vaeac_get_extra_para_default for a description of
the possible extra parameters to the vaeac approach. The
main parameters are entered directly into the explain()
function, while the extra parameters are included in a named list called
vaeac.extra_parameters.
Code Examples
We now demonstrate the vaeac approach on several
different use cases. Note that this vignette runs on CPU, but all code
sections below can be run on GPU too. To enable GPU, we have to include
vaeac.extra_parameters = list(vaeac.cuda = TRUE) in the
calls to the explain() function.
Basic Example
Here we go through how to use the vaeac approach on the
same data as in the general usage
First we load the shapr package
First we set up the model we want to explain.
library(xgboost)
library(data.table)
data("airquality")
data <- data.table::as.data.table(airquality)
data <- data[complete.cases(data), ]
x_var <- c("Solar.R", "Wind", "Temp", "Month")
y_var <- "Ozone"
ind_x_explain <- 1:6
x_train <- data[-ind_x_explain, ..x_var]
y_train <- data[-ind_x_explain, get(y_var)]
x_explain <- data[ind_x_explain, ..x_var]
# Fitting a basic xgboost model to the training data
model <- xgboost(
x = x_train,
y = y_train,
nround = 100,
verbosity = 0
)
# Specifying the phi_0, i.e. the expected prediction without any features
phi0 <- mean(y_train)First vaeac example
We are now going to explain predictions made by the model using the
vaeac approach.
n_MC_samples <- 25 # Low number of MC samples to make the vignette build faster
vaeac.n_vaeacs_initialize <- 2 # Initialize several vaeacs to counteract bad initialization values
vaeac.epochs <- 4 # The number of training epochs
explanation <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = n_MC_samples,
vaeac.epochs = vaeac.epochs,
vaeac.n_vaeacs_initialize = vaeac.n_vaeacs_initialize
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:39:21
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: vaeac
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 25
#>
#> • Number of feature-wise Shapley values: 4
#>
#> • Number of observations to explain: 6
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164830043a61.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 16 of 16 coalitions.We can look at the Shapley values.
# Printing and plotting the Shapley values.
# See ?shapr::explain for interpretation of the values.
print(explanation$shapley_values_est)
#> explain_id none Solar.R Wind Temp Month
#> <int> <num> <num> <num> <num> <num>
#> 1: 1 43.086 0.21480 -2.6541 -13.408 0.88554
#> 2: 2 43.086 4.32749 -10.8990 -17.254 -2.49949
#> 3: 3 43.086 0.27318 -11.1689 -16.801 0.59274
#> 4: 4 43.086 6.25300 -20.6611 -6.111 -2.30911
#> 5: 5 43.086 1.03750 -9.5855 -11.375 -1.64989
#> 6: 6 43.086 1.65562 -15.6976 -16.365 2.72976
plot(explanation)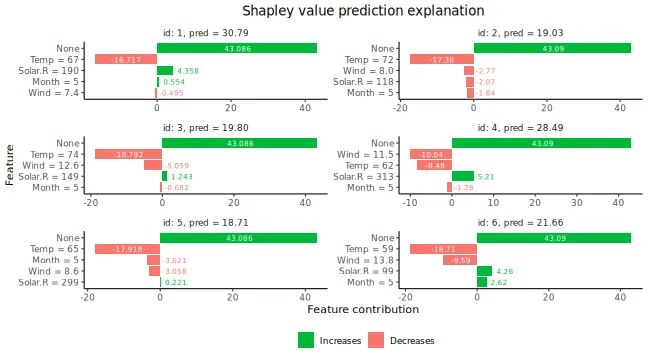
Pre-trained vaeac
If the user has a pre-trained vaeac model (from a
previous run), the user can send that to the explain()
function and shapr will skip the training of a new
vaeac model and rather use the provided vaeac
model. This is useful if we want to explain new predictions using the
same combinations/coalitions as previously, i.e., we have a new
x_explain. Note that the new x_explain must
have the same features as before.
The vaeac model is accessible via
explanation$internal$parameters$vaeac. Note that if we let
'vS_details' %in% verbose in explain(), then
shapr will give a message that it loads a pretrained
vaeac model instead of training it from scratch.
In this example, we extract the trained vaeac model from
the previous example and send it to explain().
# Send the pre-trained vaeac model
expl_pretrained_vaeac <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = n_MC_samples,
vaeac.extra_parameters = list(
vaeac.pretrained_vaeac_model = explanation$internal$parameters$vaeac
)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:39:27
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: vaeac
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 25
#>
#> • Number of feature-wise Shapley values: 4
#>
#> • Number of observations to explain: 6
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16481b8a9655.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 16 of 16 coalitions.
# Check that this version provides the same Shapley values
all.equal(explanation$shapley_values_est, expl_pretrained_vaeac$shapley_values_est)
#> [1] TRUEPre-trained vaeac (path)
We can also just provide a path to the stored vaeac
model. This is beneficial if we have only stored the vaeac
model on the computer but not the whole explanation object.
The possible save paths are stored in
explanation$internal$parameters$vaeac$model. Note that if
we let 'vS_details' %in% verbose in explain(),
then shapr will give a message that it loads a pretrained
vaeac model instead of training it from scratch.
# Call `explanation$internal$parameters$vaeac$model` to see possible vaeac models. We use `best` below.
# Send the pre-trained vaeac path
expl_pretrained_vaeac_path <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = n_MC_samples,
vaeac.extra_parameters = list(
vaeac.pretrained_vaeac_model = explanation$internal$parameters$vaeac$models$best
)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:39:32
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: vaeac
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 25
#>
#> • Number of feature-wise Shapley values: 4
#>
#> • Number of observations to explain: 6
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16483ff17d3d.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 16 of 16 coalitions.
# Check that this version provides the same Shapley values
all.equal(explanation$shapley_values_est, expl_pretrained_vaeac_path$shapley_values_est)
#> [1] TRUESpecified max_n_coalitions
In this section, we discuss a general shapr parameter in
the explain() function that is method independent, namely,
max_n_coalitions. The user can limit the Shapley value
computations to only a subset of coalitions by setting the
max_n_coalitions parameter to a value lower than
.
Note that we do not need to train a new vaeac model as
we can use the one above trained on all 16 coalitions as we
are now only using a subset of them. This is not applicable the other
way around.
# Send the pre-trained vaeac path
expl_batches_coalitions <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
max_n_coalitions = 10,
n_MC_samples = n_MC_samples,
vaeac.extra_parameters = list(
vaeac.pretrained_vaeac_model = explanation$internal$parameters$vaeac
)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:39:36
#>
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: vaeac
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 25
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 6
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164813e89b6b.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 10 of 16 coalitions.
# Gives different Shapley values as the latter are based only on a subset of coalitions
plot_SV_several_approaches(list("Original" = explanation, "Other coals." = expl_batches_coalitions))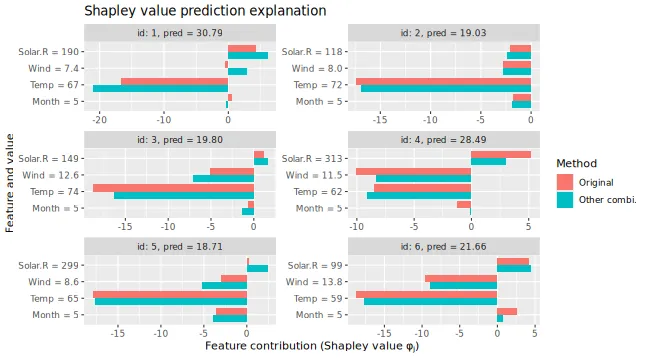
# Can compare that to the situation where we have exact computations (i.e., include all coalitions)
explanation$internal$objects$X
#> Index: <coalition_size>
#> id_coalition coalitions coalitions_str coalition_size N shapley_weight
#> <int> <list> <char> <int> <int> <num>
#> 1: 1 0 1 1.00e+06
#> 2: 2 1 1 1 4 2.50e-01
#> 3: 3 2 2 1 4 2.50e-01
#> 4: 4 3 3 1 4 2.50e-01
#> 5: 5 4 4 1 4 2.50e-01
#> 6: 6 1,2 1 2 2 6 1.25e-01
#> 7: 7 1,3 1 3 2 6 1.25e-01
#> 8: 8 1,4 1 4 2 6 1.25e-01
#> 9: 9 2,3 2 3 2 6 1.25e-01
#> 10: 10 2,4 2 4 2 6 1.25e-01
#> 11: 11 3,4 3 4 2 6 1.25e-01
#> 12: 12 1,2,3 1 2 3 3 4 2.50e-01
#> 13: 13 1,2,4 1 2 4 3 4 2.50e-01
#> 14: 14 1,3,4 1 3 4 3 4 2.50e-01
#> 15: 15 2,3,4 2 3 4 3 4 2.50e-01
#> 16: 16 1,2,3,4 1 2 3 4 4 1 1.00e+06
#> sample_freq features approach
#> <lgcl> <list> <char>
#> 1: NA <NA>
#> 2: NA 1 vaeac
#> 3: NA 2 vaeac
#> 4: NA 3 vaeac
#> 5: NA 4 vaeac
#> 6: NA 1,2 vaeac
#> 7: NA 1,3 vaeac
#> 8: NA 1,4 vaeac
#> 9: NA 2,3 vaeac
#> 10: NA 2,4 vaeac
#> 11: NA 3,4 vaeac
#> 12: NA 1,2,3 vaeac
#> 13: NA 1,2,4 vaeac
#> 14: NA 1,3,4 vaeac
#> 15: NA 2,3,4 vaeac
#> 16: NA 1,2,3,4 <NA>Note that if we train a vaeac model from scratch with
the setup above, then the vaeac model will not use an MCAR
(missing completely at random) mask generator, but rather a mask
generator that ensures that the vaeac model is only trained
on the specified set of coalitions. In this case, it will be the set of
the sampled coalitions.
expl_batches_coalitions_2 <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
max_n_coalitions = 10,
n_MC_samples = n_MC_samples,
vaeac.n_vaeacs_initialize = 1,
vaeac.epochs = 3,
verbose = "vS_details"
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:39:40
#>
#> ── Extra info about the pretrained vaeac model ──
#>
#> Training the `vaeac` model with the provided
#> parameters from scratch on CPU.
#> Using 'specified_masks_mask_generator' with '8'
#> coalitions.
#> The vaeac model contains 17032 trainable parameters.
#> Initializing vaeac model number 1 of 1.
#> Best vaeac inititalization was number 1 (of 1) with a
#> training VLB = -6.49 after 2 epochs. Continue to
#> train this inititalization.
#>
#> Results of the `vaeac` training process:
#> Best epoch: 3. VLB = -4.812 IWAE = -3.235 IWAE_running = -3.542
#> Best running avg epoch: 3. VLB = -4.812 IWAE = -3.235 IWAE_running = -3.542
#> Last epoch: 3. VLB = -4.812 IWAE = -3.235 IWAE_running = -3.542
#> ℹ The trained `vaeac` models are saved to folder '/tmp/Rtmpravdkd' at
#> '/tmp/Rtmpravdkd/X2026.01.16.12.39.40.368448_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.39.40.368448_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best_running.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.39.40.368448_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_last.pt'Paired sampling
The vaeac approach can use paired sampling to improve
the stability of the vaeac training procedure. When using paired
sampling, each observation in the training batches will be duplicated,
but the first version will be masked by
and the second version will be masked by the complement
.
The masks are taken from the explanation$internal$objects$S
matrix. Note that vaeac does not check if the complement is
also in said matrix. This means that if the Shapley value explanations
are computed based on a subset of coalitions, then the
vaeac model might be trained on coalitions which are not
used when computing the Shapley values. This should not be considered as
redundant training as it increases the stability and performance of the
vaeac model as a whole; hence, we recommend using paired
sampling (default). Furthermore, the masks are randomly selected for
each observation in the batch. The training time when using paired
sampling is higher in comparison to random sampling due to more complex
implementation.
expl_paired_sampling_TRUE <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = n_MC_samples,
vaeac.epochs = 10,
vaeac.n_vaeacs_initialize = 1,
vaeac.extra_parameters = list(vaeac.paired_sampling = TRUE)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:39:46
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: vaeac
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 25
#>
#> • Number of feature-wise Shapley values: 4
#>
#> • Number of observations to explain: 6
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16487261121c.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 16 of 16 coalitions.
expl_paired_sampling_FALSE <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = n_MC_samples,
vaeac.epochs = 10,
vaeac.n_vaeacs_initialize = 1,
vaeac.extra_parameters = list(vaeac.paired_sampling = FALSE)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:39:56
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: vaeac
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 25
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 6
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16482db56d4b.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.We can compare the results by looking at the training and validation
errors and by the
evaluation criterion. We do this by using the
plot_vaeac_eval_crit() and
plot_MSEv_eval_crit() functions in the shapr
package, respectively.
explanation_list <- list("Regular samp." = expl_paired_sampling_FALSE,
"Paired samp." = expl_paired_sampling_TRUE)
plot_vaeac_eval_crit(explanation_list, plot_type = "criterion")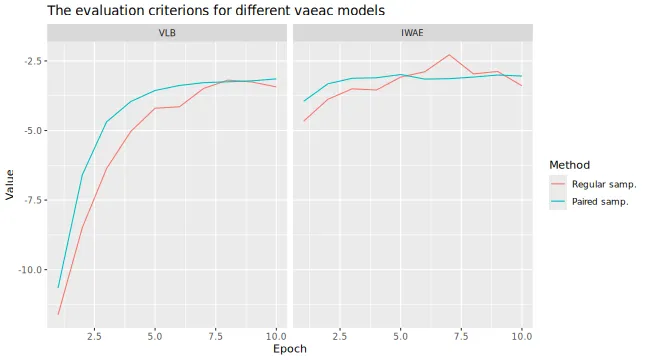
plot_MSEv_eval_crit(explanation_list)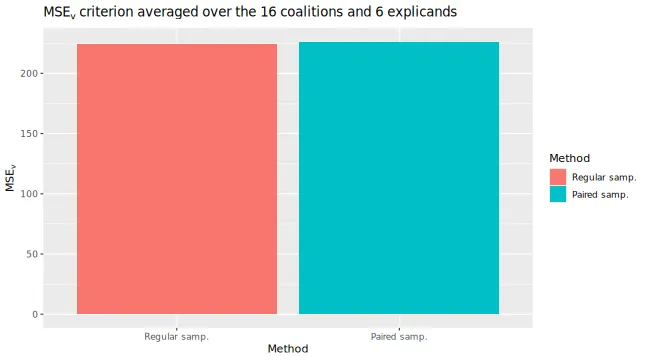
By looking at the time, we see that the paired version takes (a bit)
longer time in the setup_computation phase, that is, in the
training phase.
rbind(
"Paired" = expl_paired_sampling_TRUE$timing$overall_timing_secs,
"Regular" = expl_paired_sampling_FALSE$timing$overall_timing_secs
)
#> setup test_prediction main_computation finalize_explanation
#> Paired 0.048663 0.076673 9.4586 0.0079832
#> Regular 0.065304 0.049553 7.0216 0.0037577Progressr
As discussed in the general usage, the shapr package
provides two ways for receiving information about the progress of the
approach. First, the shapr package provides progress
updates of the computation of the Shapley values through the
progressr package. Second, the user can also get various
form of information through verbose in
explain(). By letting
'vS_details' %in% verbose, we get extra information related
to the vaeac approach. The verbose parameter
works independently of the progressr package. Meaning that
the user can chose to use none, either, or both options simultaneously.
We give two examples here, and refer the reader to the general usage for
more detailed information.
By setting c("basic", "vS_details"), we get both basic
messages about the explanation case and messages about the estimation of
the vaeac approach.
expl_with_messages <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = n_MC_samples,
verbose = c("basic","vS_details"),
vaeac.epochs = 5,
vaeac.n_vaeacs_initialize = 2
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:40:05
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: vaeac
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 25
#>
#> • Number of feature-wise Shapley values: 4
#>
#> • Number of observations to explain: 6
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164823ec31fb.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 16 of 16 coalitions.
#>
#>
#>
#> ── Extra info about the pretrained vaeac model ──
#>
#>
#>
#> Training the `vaeac` model with the provided
#> parameters from scratch on CPU.
#>
#> Using 'mcar_mask_generator' with 'masking_ratio =
#> 0.5'.
#>
#> The vaeac model contains 17032 trainable parameters.
#>
#> Initializing vaeac model number 1 of 2.
#>
#> Initializing vaeac model number 2 of 2.
#>
#> Best vaeac inititalization was number 2 (of 2) with a
#> training VLB = -4.566 after 2 epochs. Continue to
#> train this inititalization.
#>
#>
#> Results of the `vaeac` training process:
#> Best epoch: 5. VLB = -3.318 IWAE = -3.049 IWAE_running = -3.149
#> Best running avg epoch: 5. VLB = -3.318 IWAE = -3.049 IWAE_running = -3.149
#> Last epoch: 5. VLB = -3.318 IWAE = -3.049 IWAE_running = -3.149
#>
#> ℹ The trained `vaeac` models are saved to folder '/tmp/Rtmpravdkd' at
#> '/tmp/Rtmpravdkd/X2026.01.16.12.40.05.495272_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.40.05.495272_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best_running.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.40.05.495272_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_last.pt'For more visual information we can use the progressr
package. This can help us see detailed progress of the training step for
the final vaeac model. Note that by default
vS_details is not part of verbose, meaning
that we do not get any messages from the vaeac approach and
only get the progress bars. See the general usage for examples for how
to change the progress bar.
library(progressr)
progressr::handlers("cli") # Use `progressr::handlers("void")` to silence all `progressr` updates
progressr::with_progress({
expl_with_progressr <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = n_MC_samples,
verbose = "vS_details",
vaeac.epochs = 5,
vaeac.n_vaeacs_initialize = 2
)
})
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:40:15
#>
#> ── Extra info about the pretrained vaeac model ──
#>
#> Training the `vaeac` model with the provided
#> parameters from scratch on CPU.
#> Using 'mcar_mask_generator' with 'masking_ratio =
#> 0.5'.
#> The vaeac model contains 17032 trainable parameters.
#> Initializing vaeac model number 1 of 2.
#>
[KInitializing vaeac model number 2 of 2.
#> ■■■■■■■■■■ 29% | Training vaea…
[KBest vaeac inititalization was number 2 (of 2) with a
#> training VLB = -4.566 after 2 epochs. Continue to
#> train this inititalization.
#> ■■■■■■■■■■■■■■■■■■ 57% | Training vaea…
#> Results of the `vaeac` training process:
#> Best epoch: 5. VLB = -3.318 IWAE = -3.049 IWAE_running = -3.149
#> Best running avg epoch: 5. VLB = -3.318 IWAE = -3.049 IWAE_running = -3.149
#> Last epoch: 5. VLB = -3.318 IWAE = -3.049 IWAE_running = -3.149
#> ℹ The trained `vaeac` models are saved to folder '/tmp/Rtmpravdkd' at
#> '/tmp/Rtmpravdkd/X2026.01.16.12.40.14.973098_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.40.14.973098_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best_running.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.40.14.973098_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_last.pt'
all.equal(expl_with_messages$shapley_values_est, expl_with_progressr$shapley_values_est)
#> [1] TRUEContinue the training of the vaeac approach
In the case the user has set a too low number of training epochs and
sees that the network is still learning, then the user can continue to
train the network from where it stopped. Thus, a good workflow can
therefore be to call the explain() function with
n_MC_samples = 1 (to avoid spending too much time
generating MC samples), then look at the training and evaluation plots
of the vaeac. If not satisfied, then train more. If
satisfied, then call the explain() function again, but this
time using the extra parameter
vaeac.pretrained_vaeac_model, as illustrated above. Note
that we have set the number of vaeac.epochs to be very low
in this example and we recommend using many more epochs.
We can compare the results by looking at the training and validation
errors and by the
evaluation criterion. We do this by using the
plot_vaeac_eval_crit() and
plot_MSEv_eval_crit() functions in the shapr
package, respectively. We also use the
plot_vaeac_imputed_ggpairs() function which generates
samples from
,
this is meant as a sanity check to see that the vaeac model
is able to follow the general structure/distribution of the data.
However, recall that the vaeac model is never trained on
the empty coalition, so the produced samples should be taken with a
grain of salt.
expl_little_training <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = 250,
vaeac.epochs = 3,
vaeac.n_vaeacs_initialize = 2
)
# Look at the training and validation errors. Not happy and want to train more.
plot_vaeac_eval_crit(list("Original" = expl_little_training), plot_type = "method")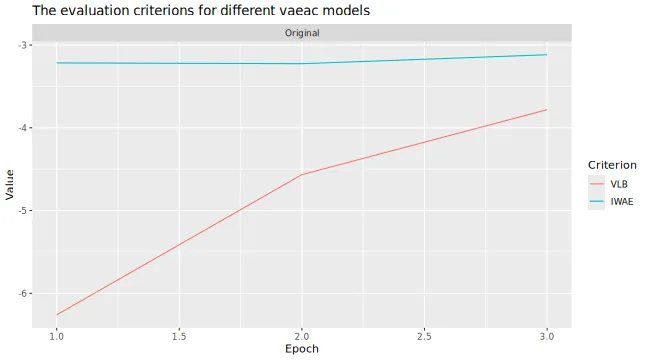
# Can also see how well vaeac generates data from the full joint distribution. Quite good.
plot_vaeac_imputed_ggpairs(
explanation = expl_little_training,
which_vaeac_model = "best",
x_true = x_train
) + ggplot2::labs(title = NULL)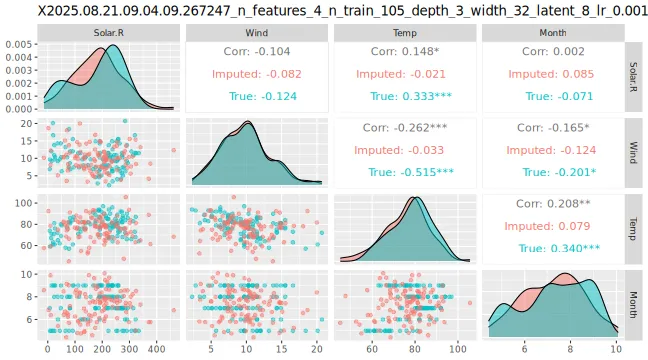
# Make a copy of the explanation object and continue to train the vaeac model some more epochs
expl_train_more <- expl_little_training
expl_train_more$internal$parameters$vaeac <-
vaeac_train_model_continue(
explanation = expl_train_more,
epochs_new = 5,
x_train = x_train
)
# Compute the Shapley values again but this time using the extra trained vaeac model
expl_train_more_vaeac <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = 250,
vaeac.extra_parameters = list(
vaeac.pretrained_vaeac_model = expl_train_more$internal$parameters$vaeac
)
)
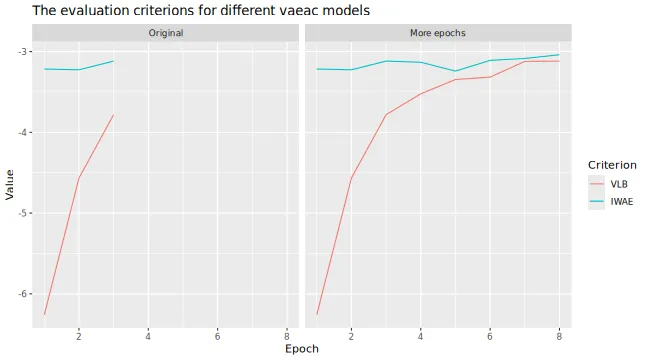
# Look at the training and validation errors and conclude that we want to train some more
plot_vaeac_eval_crit(
list("Original" = expl_little_training, "More epochs" = expl_train_more),
plot_type = "method"
)
# Continue to train the vaeac model some more epochs
expl_train_even_more <- expl_train_more
expl_train_even_more$internal$parameters$vaeac <-
vaeac_train_model_continue(
explanation = expl_train_even_more,
epochs_new = 10,
x_train = x_train
)
# Compute the Shapley values again but this time using the even more trained vaeac model
expl_train_even_more_vaeac <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = 250,
vaeac.extra_parameters = list(
vaeac.pretrained_vaeac_model = expl_train_even_more$internal$parameters$vaeac
)
)
# Look at the training and validation errors.
plot_vaeac_eval_crit(
list(
"Original" = expl_little_training,
"More epochs" = expl_train_more,
"Even more epochs" = expl_train_even_more
),
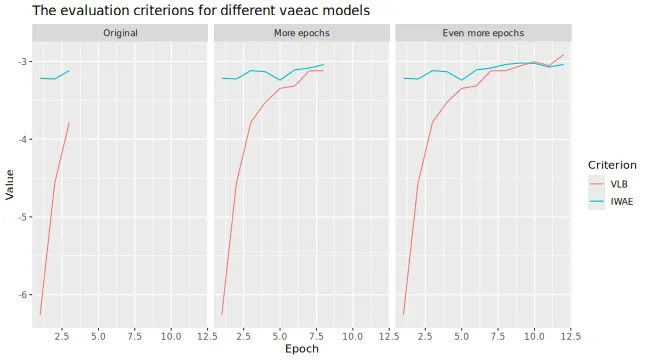
plot_type = "method"
)
# Can also see how well vaeac generates data from the full joint distribution
plot_vaeac_imputed_ggpairs(
explanation = expl_train_even_more,
which_vaeac_model = "best",
x_true = x_train
) + ggplot2::labs(title = NULL)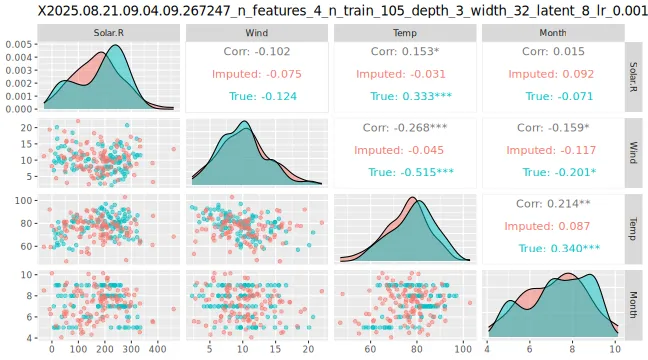
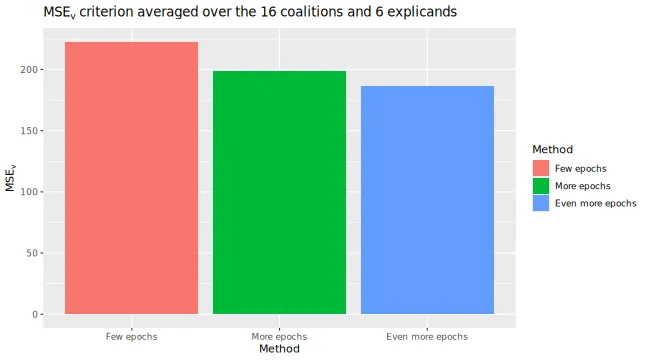
We can see that the extra training has decreased the MSEv score. The Shapley value explanations have also changed, but they are often comparable.
plot_MSEv_eval_crit(list(
"Few epochs" = expl_little_training,
"More epochs" = expl_train_more_vaeac,
"Even more epochs" = expl_train_even_more_vaeac
))
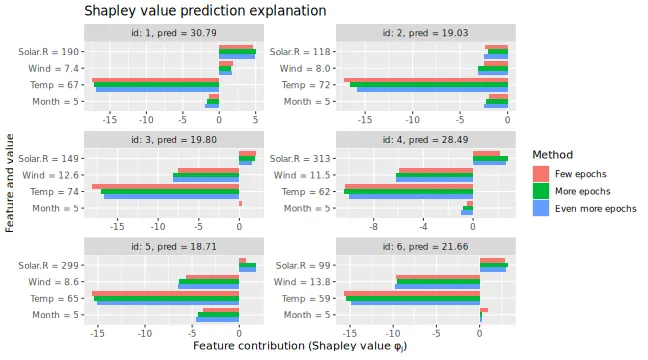
# We see that the Shapley values have changed, but they are often comparable
plot_SV_several_approaches(list(
"Few epochs" = expl_little_training,
"More epochs" = expl_train_more_vaeac,
"Even more epochs" = expl_train_even_more_vaeac
))
Vaeac with early stopping
If we do not want to specify the number of epochs, as we
are uncertain how many epochs it will take before the
vaeac model is properly trained, a good choice is to rather
use early stopping. This means that we can set vaeac.epochs
to a large number and let vaeac.epochs_early_stopping be
for example 5. This means that the vaeac model
will stop the training procedure if there has been no improvement in the
validation score for 5 epochs.
# Low value for `vaeac.epochs_early_stopping` here to build the vignette faster
expl_early_stopping <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = 250,
verbose = c("basic","vS_details"),
vaeac.epochs = 1000, # Set it to a big number
vaeac.n_vaeacs_initialize = 2,
vaeac.extra_parameters = list(vaeac.epochs_early_stopping = 2)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:41:55
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: vaeac
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 250
#>
#> • Number of feature-wise Shapley values: 4
#>
#> • Number of observations to explain: 6
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16484845e94.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 16 of 16 coalitions.
#>
#>
#>
#> ── Extra info about the pretrained vaeac model ──
#>
#>
#>
#> Training the `vaeac` model with the provided
#> parameters from scratch on CPU.
#>
#> Using 'mcar_mask_generator' with 'masking_ratio =
#> 0.5'.
#>
#> The vaeac model contains 17032 trainable parameters.
#>
#> Initializing vaeac model number 1 of 2.
#>
#> Initializing vaeac model number 2 of 2.
#>
#> Best vaeac inititalization was number 2 (of 2) with a
#> training VLB = -4.566 after 2 epochs. Continue to
#> train this inititalization.
#>
#> No IWAE improvment in 2 epochs. Apply early stopping
#> at epoch 14.
#>
#>
#> Results of the `vaeac` training process:
#> Best epoch: 12. VLB = -2.958 IWAE = -2.930 IWAE_running = -2.991
#> Best running avg epoch: 12. VLB = -2.958 IWAE = -2.930 IWAE_running = -2.991
#> Last epoch: 14. VLB = -2.971 IWAE = -2.955 IWAE_running = -2.996
#>
#> ℹ The trained `vaeac` models are saved to folder '/tmp/Rtmpravdkd' at
#> '/tmp/Rtmpravdkd/X2026.01.16.12.41.54.927568_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.41.54.927568_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best_running.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.41.54.927568_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_last.pt'
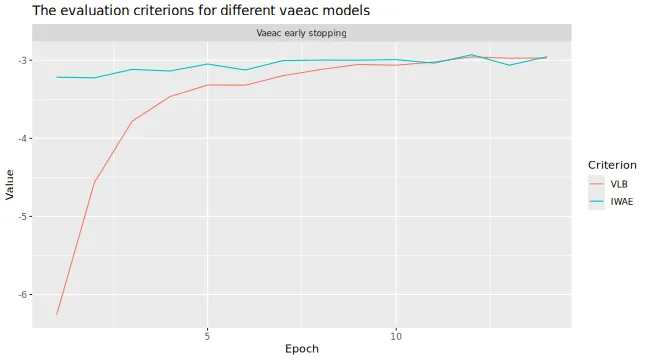
# Look at the training and validation errors. We are quite happy with it.
plot_vaeac_eval_crit(
list("Vaeac early stopping" = expl_early_stopping),
plot_type = "method"
)
However, we can train it further for a fixed amount of epochs if
desired. This can be in a setting where we are not happy with the IWAE
curve or we feel that we set vaeac.epochs_early_stopping to
a too low value or if the max number of epochs
(vaeac.epochs) were reached.
# Make a copy of the explanation object which we are to train further.
expl_early_stopping_train_more <- expl_early_stopping
# Continue to train the vaeac model some more epochs
expl_early_stopping_train_more$internal$parameters$vaeac <-
vaeac_train_model_continue(
explanation = expl_early_stopping_train_more,
epochs_new = 15,
x_train = x_train,
verbose = NULL
)
# Can even do it twice if desired
expl_early_stopping_train_more$internal$parameters$vaeac <-
vaeac_train_model_continue(
explanation = expl_early_stopping_train_more,
epochs_new = 10,
x_train = x_train,
verbose = NULL
)
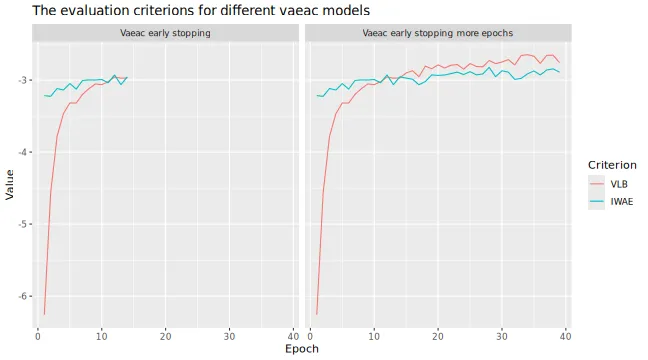
# Look at the training and validation errors. We see some improvement
plot_vaeac_eval_crit(
list(
"Vaeac early stopping" = expl_early_stopping,
"Vaeac early stopping more epochs" = expl_early_stopping_train_more
),
plot_type = "method"
)
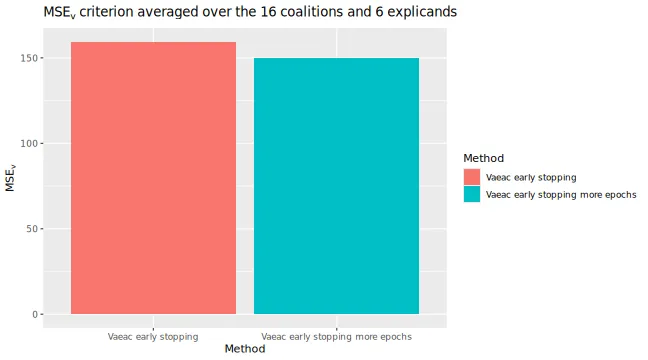
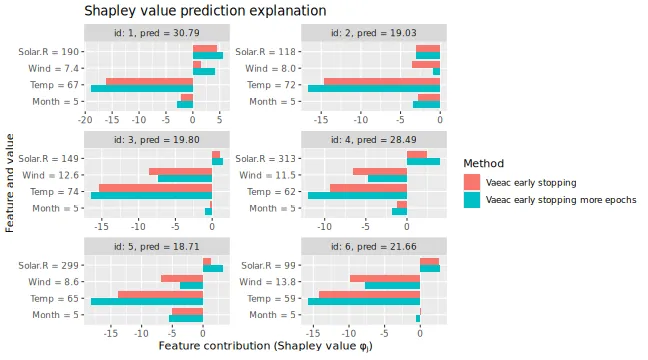
We can then use the extra trained version to compute the Shapley value explanations and compare it with the previous version that used early stopping. We see a non-significant difference.
# Use extra trained vaeac model to compute Shapley values again.
expl_early_stopping_train_more <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = 250,
vaeac.extra_parameters = list(
vaeac.pretrained_vaeac_model = expl_early_stopping_train_more$internal$parameters$vaeac
)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:42:51
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: vaeac
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 250
#>
#> • Number of feature-wise Shapley values: 4
#>
#> • Number of observations to explain: 6
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16481d393199.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 16 of 16 coalitions.
# We can compare their MSEv scores
plot_MSEv_eval_crit(list(
"Vaeac early stopping" = expl_early_stopping,
"Vaeac early stopping more epochs" = expl_early_stopping_train_more
))
# We see that the Shapley values have changed, but only slightly
plot_SV_several_approaches(list(
"Vaeac early stopping" = expl_early_stopping,
"Vaeac early stopping more epochs" = expl_early_stopping_train_more
))
Grouping of features
When we train a vaeac model to explain groups of
features, then the vaeac model will use the
“Specified_masks_mask_generator”, which ensures that the
vaeac model only trains on a specified set of coalitions.
In this case, it will ensure that all features in group A will always
either be conditioned on or be unconditioned. The same goes for group B.
Note that in this setup, there are only 4 possible
coalitions, but vaeac only trains on 2
coalitions as the empty and grand coalitions as they are not needed in
the Shapley value computations.
expl_group <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "vaeac",
phi0 = phi0,
seed = 1,
group = list(A = c("Temp", "Month"), B = c("Wind", "Solar.R")),
n_MC_samples = n_MC_samples,
verbose = "vS_details",
vaeac.epochs = 4,
vaeac.n_vaeacs_initialize = 2
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:43:14
#>
#> ── Extra info about the pretrained vaeac model ──
#>
#> Training the `vaeac` model with the provided
#> parameters from scratch on CPU.
#> Using 'specified_masks_mask_generator' with '2'
#> coalitions.
#> The vaeac model contains 17032 trainable parameters.
#> Initializing vaeac model number 1 of 2.
#> Initializing vaeac model number 2 of 2.
#> Best vaeac inititalization was number 2 (of 2) with a
#> training VLB = -4.814 after 2 epochs. Continue to
#> train this inititalization.
#>
#> Results of the `vaeac` training process:
#> Best epoch: 3. VLB = -3.935 IWAE = -3.124 IWAE_running = -3.267
#> Best running avg epoch: 4. VLB = -3.619 IWAE = -3.138 IWAE_running = -3.235
#> Last epoch: 4. VLB = -3.619 IWAE = -3.138 IWAE_running = -3.235
#> ℹ The trained `vaeac` models are saved to folder '/tmp/Rtmpravdkd' at
#> '/tmp/Rtmpravdkd/X2026.01.16.12.43.13.96921_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.43.13.96921_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_best_running.pt'
#> '/tmp/Rtmpravdkd/X2026.01.16.12.43.13.96921_n_features_4_n_train_105_depth_3_width_32_latent_8_lr_0.001_epoch_last.pt'
# Plot the resulting explanations
plot(expl_group)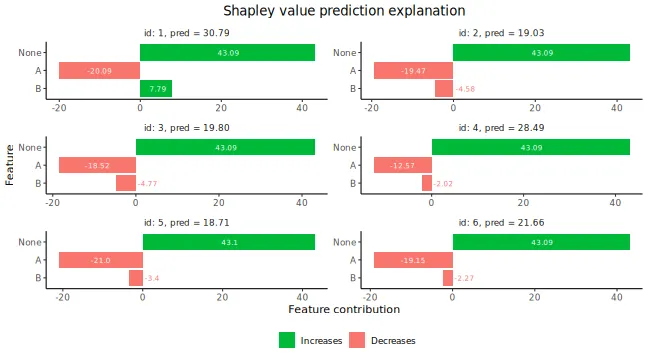
Mixed Data
Here we look at a setup with mixed data, i.e., the data contains both categorical and continuous features. First we set up the data and the model.
library(ranger)
#> ranger 0.17.0 using 2 threads (default). Change with num.threads in ranger() and predict(), options(Ncpus = N), options(ranger.num.threads = N) or environment variable R_RANGER_NUM_THREADS.
data <- data.table::as.data.table(airquality)
data <- data[complete.cases(data), ]
# Convert the month variable to a factor
data[, Month_factor := as.factor(Month)]
x_var_cat <- c("Solar.R", "Wind", "Temp", "Month_factor")
y_var <- "Ozone"
ind_x_explain <- 1:6
data_train_cat <- data[-ind_x_explain, ]
x_train_cat <- data_train_cat[, ..x_var_cat]
x_explain_cat <- data[ind_x_explain, ][, ..x_var_cat]
# Fit a random forest model to the training data
model <- ranger(as.formula(paste0(y_var, " ~ ", paste0(x_var_cat, collapse = " + "))),
data = data_train_cat
)
# Specifying the phi_0, i.e. the expected prediction without any features
phi0 <- mean(data_train_cat[, get(y_var)])Then we compute explanations using the ctree and
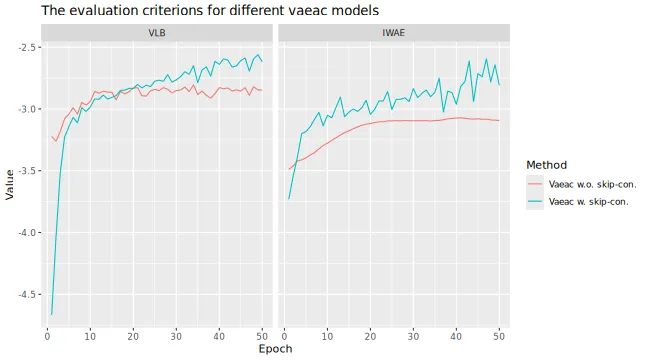
vaeac approaches. For the vaeac approach, we
consider two setups: the default architecture, and a simpler one without
skip connections. We do this to illustrate that the skip connections
improve the vaeac method. We use ctree with
default parameters.
# Here we use the ctree approach
expl_ctree <- explain(
model = model,
x_explain = x_explain_cat,
x_train = x_train_cat,
approach = "ctree",
phi0 = phi0,
seed = 1,
n_MC_samples = 250
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:43:21
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <ranger>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: ctree
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 250
#>
#> • Number of feature-wise Shapley values: 4
#>
#> • Number of observations to explain: 6
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164826e36431.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 16 of 16 coalitions.
# Then we use the vaeac approach
expl_vaeac_with <- explain(
model = model,
x_explain = x_explain_cat,
x_train = x_train_cat,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = 250,
vaeac.epochs = 50,
vaeac.n_vaeacs_initialize = 4
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:43:23
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#> ── Explanation overview ──
#>
#> • Model class: <ranger>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: vaeac
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 250
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 6
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164867df80c7.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
# Then we use the vaeac approach
expl_vaeac_without <- explain(
model = model,
x_explain = x_explain_cat,
x_train = x_train_cat,
approach = "vaeac",
phi0 = phi0,
seed = 1,
n_MC_samples = 250,
vaeac.epochs = 50,
vaeac.n_vaeacs_initialize = 4,
vaeac.extra_parameters = list(
vaeac.skip_conn_layer = FALSE,
vaeac.skip_conn_masked_enc_dec = FALSE
)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:44:23
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 16`, and is therefore set to
#> `2^n_features = 16`.
#> ── Explanation overview ──
#>
#> • Model class: <ranger>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: vaeac
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 250
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 6
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16487250e88.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
# We see that the `vaeac` model without the skip connections performs worse
plot_vaeac_eval_crit(
list(
"Vaeac w.o. skip-con." = expl_vaeac_without,
"Vaeac w. skip-con." = expl_vaeac_with
),
plot_type = "criterion"
)
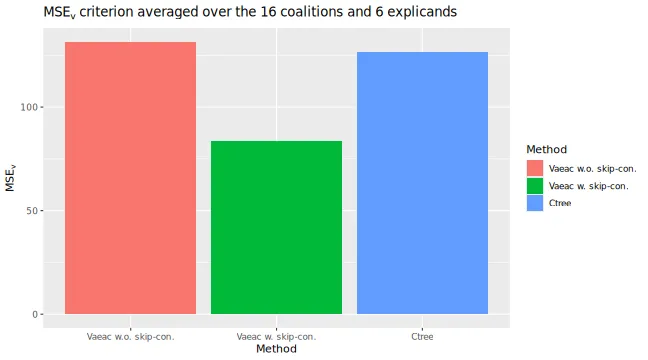
# The `vaeac` model with skip connections has the lowest/best MSE_Frye evaluation criterion score
plot_MSEv_eval_crit(list(
"Vaeac w.o. skip-con." = expl_vaeac_without,
"Vaeac w. skip-con." = expl_vaeac_with,
"Ctree" = expl_ctree
))
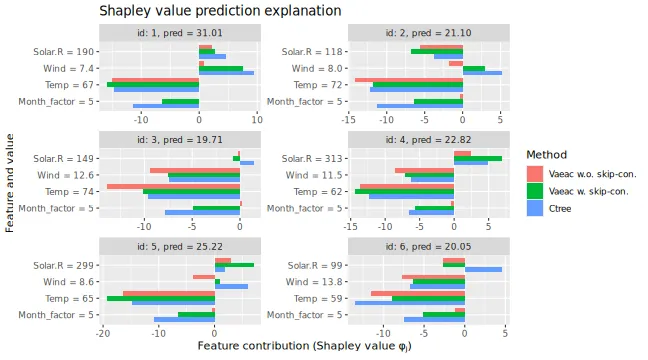
# Can compare the Shapley values. Ctree and vaeac with skip connections produce similar explanations.
plot_SV_several_approaches(
list(
"Vaeac w.o. skip-con." = expl_vaeac_without,
"Vaeac w. skip-con." = expl_vaeac_with,
"Ctree" = expl_ctree
),
index_explicands = 1:6
)