Asymmetric and causal Shapley value explanations
Lars Henry Berge Olsen
Source:vignettes/asymmetric_causal.Rmd
asymmetric_causal.RmdOverview
This vignette elaborates and demonstrates the asymmetric and causal
Shapley value frameworks introduced by Frye,
Rowat, and Feige (2020) and Heskes et al.
(2020), respectively. We also consider the marginal and
conditional Shapley value frameworks, see Lundberg and Lee (2017) and Aas, Jullum, and Løland (2021), respectively. We
demonstrate the frameworks on the bike sharing dataset
from the UCI Machine Learning Repository. The setup is based on the
CauSHAPley package, which is the code
supplement to the Heskes et al. (2020)
paper. The CauSHAPley package was based on an old version
of shapr and was restricted to the gaussian
approach (see section 6 in Heskes et al.
(2020) for more details).
We have extended the causal Shapley value framework to work for all
Monte Carlo-based approaches (independence (not
recommended), empirical, gaussian,
copula, ctree, vaeac and
categorical), while the extension of the asymmetric Shapley
value framework works for all the Monte Carlo and regression-based
approaches. Our generalization is of uttermost importance, as many
real-world data sets are far from the Gaussian distribution, and,
compared to CauSHAPley, our implementation can utilize all
of shapr’s new features, such as batch computation,
parallelization and iterative computation for both feature-wise and
group-wise Shapley values.
The main differences between the marginal, conditional, and causal Shapley value frameworks are that they sample/generate the Monte Carlo samples from the marginal distribution, (conventional) observational conditional distribution, and interventional conditional distribution, respectively. Asymmetric means that we do not consider all possible coalitions, but rather only the coalitions that respect a causal ordering.
Asymmetric conditional Shapley values
Asymmetric (conditional) Shapley values were proposed by Frye, Rowat, and Feige (2020) as a way to incorporate causal knowledge in the real world by computing the Shapley value explanations using only the feature combinations/coalitions consistent with a (partial) causal ordering. See the figure below for a schematic overview of the causal ordering we are going to use in the examples in this vignette. In the figure, we see that our causal ordering consists of three components: , , and . See the code section for what the features represent.
To elaborate, instead of considering the possible coalitions, where is the number of features, asymmetric Shapley values only consider the subset of coalitions which respects the causal ordering. For our causal ordering, this means that the asymmetric Shapley value explanation framework skips the coalitions where is included but , as is the ancestor of . This will skew the explanations towards distal/root causes, see Section 3.2 in Frye, Rowat, and Feige (2020).
We can use all approaches in shapr, both Monte
Carlo-based and regression-based methods, to compute the asymmetric
Shapley values. This is because the asymmetric Shapley value explanation
framework does not change how we compute the contribution functions
,
but rather which of the coalitions
that are used to compute the Shapley value explanations. This means that
the number of coalitions is no longer
,
but rather
,
where
is the number of features
()
in the largest component of the causal ordering.
Furthermore, asymmetric Shapley values support groups of features, but then the causal ordering must be given on the group level instead of on the feature level. The asymmetric Shapley value framework also supports sampling of coalitions where the sampling is done from the set of coalitions that respect the causal ordering.
Finally, we remark that asymmetric conditional Shapley values are equivalent to asymmetric causal Shapley values (see below) when we only use the coalitions respecting the causal ordering and assuming that all dependencies within chain components are induced by mutual interactions.
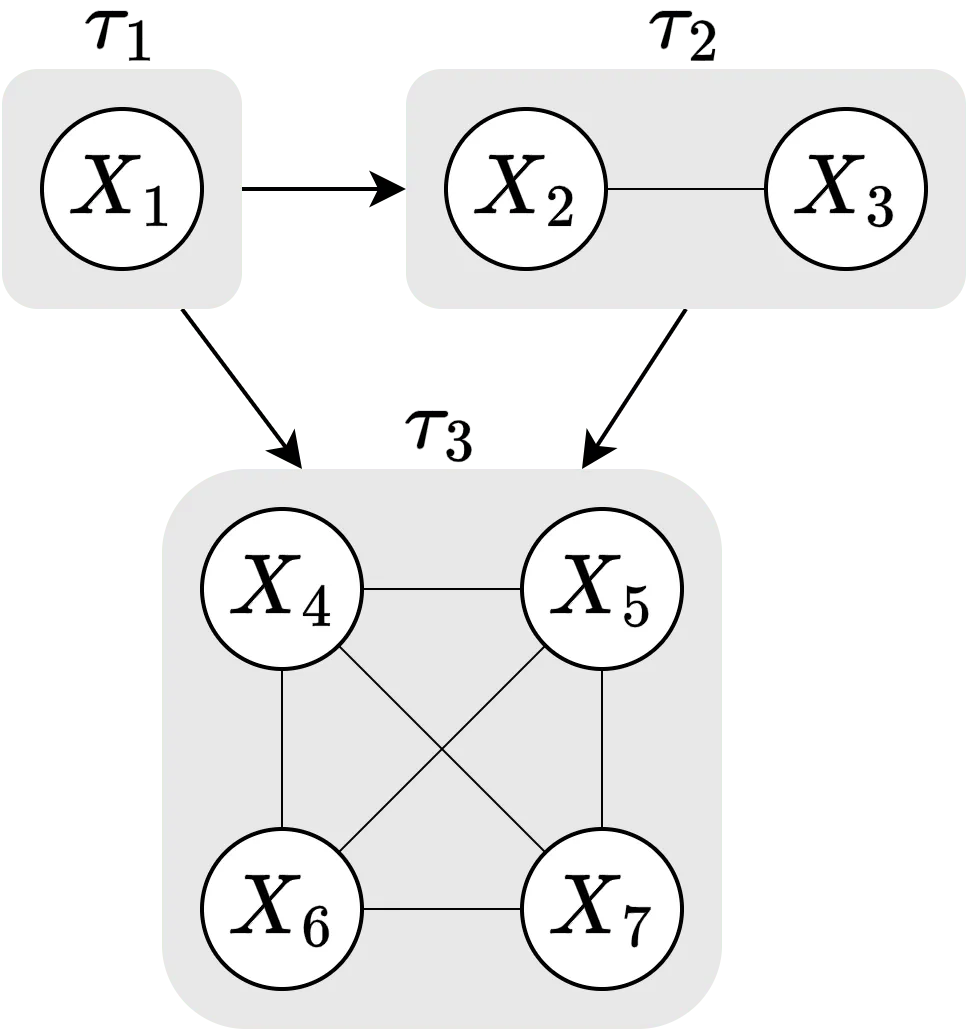
Schematic overview of the causal ordering used in this vignette.
Causal Shapley values
Causal Shapley values were proposed by Heskes
et al. (2020) as a way to explain the total effect of features on
the prediction by taking into account their causal relationships and
adapting the sampling procedure in shapr. More precisely,
they propose to employ Pearl’s do-calculus to circumvent the
independence assumption, made by Lundberg and Lee
(2017), without sacrificing any of the desirable properties of
the Shapley value framework. The causal Shapley value explanation
framework can also separate the contribution of direct and indirect
effects, which makes them principally different from marginal and
conditional Shapley values. The framework also provides a more direct
and robust way to incorporate causal knowledge, compared to the
asymmetric Shapley value explanation framework.
To compute causal Shapley values, we have to specify a (partial) causal ordering and make an assumption about the confounding in each component. Together, they form a causal chain graph which contains directed and undirected edges. All features that are treated on an equal footing are linked together with undirected edges and become part of the same chain component. Edges between chain components are directed and represent causal relationships. In the figure below, we have the same causal ordering as above, but we have in addition made the assumption that we have confounding in the second component, but no confounding in the first and third components. This allows us to correctly distinguish between dependencies that are due to confounding and mutual interactions. That is, in the figure, the dependencies in chain component are assumed to be the result of a common confounder, and those in of mutual interactions, while we have no mutual interactions in as it is a singleton.
Computing the effect of an intervention depends on how we interpret the generative process that leads to the feature dependencies within each component. If they are the result of marginalizing out a common confounder, then intervention on a particular feature will break the dependency with the other features, and we denote the set of these chain components by . For the components with mutual feature interactions, setting the value of a feature affects the distribution of the variables within the same component. We denote the set of these components by .
Heskes et al. (2020) described how any
expectation by intervention needed to compute the causal Shapley values
can be translated to an expectation by observation, by using the
interventional formula for causal chain graphs:
Here, any of the Monte
Carlo-based approaches in shapr can be used to compute the
conditional distributions/observational expectations. The marginals are
estimated from the training data for all approaches except
gaussian, for which we use the marginals of the Gaussian
distribution instead.
For specific causal chain graphs, the causal Shapley value framework simplifies to symmetric conditional, asymmetric conditional, and marginal Shapley values; see Corollaries 1 to 3 in the supplement of Heskes et al. (2020).
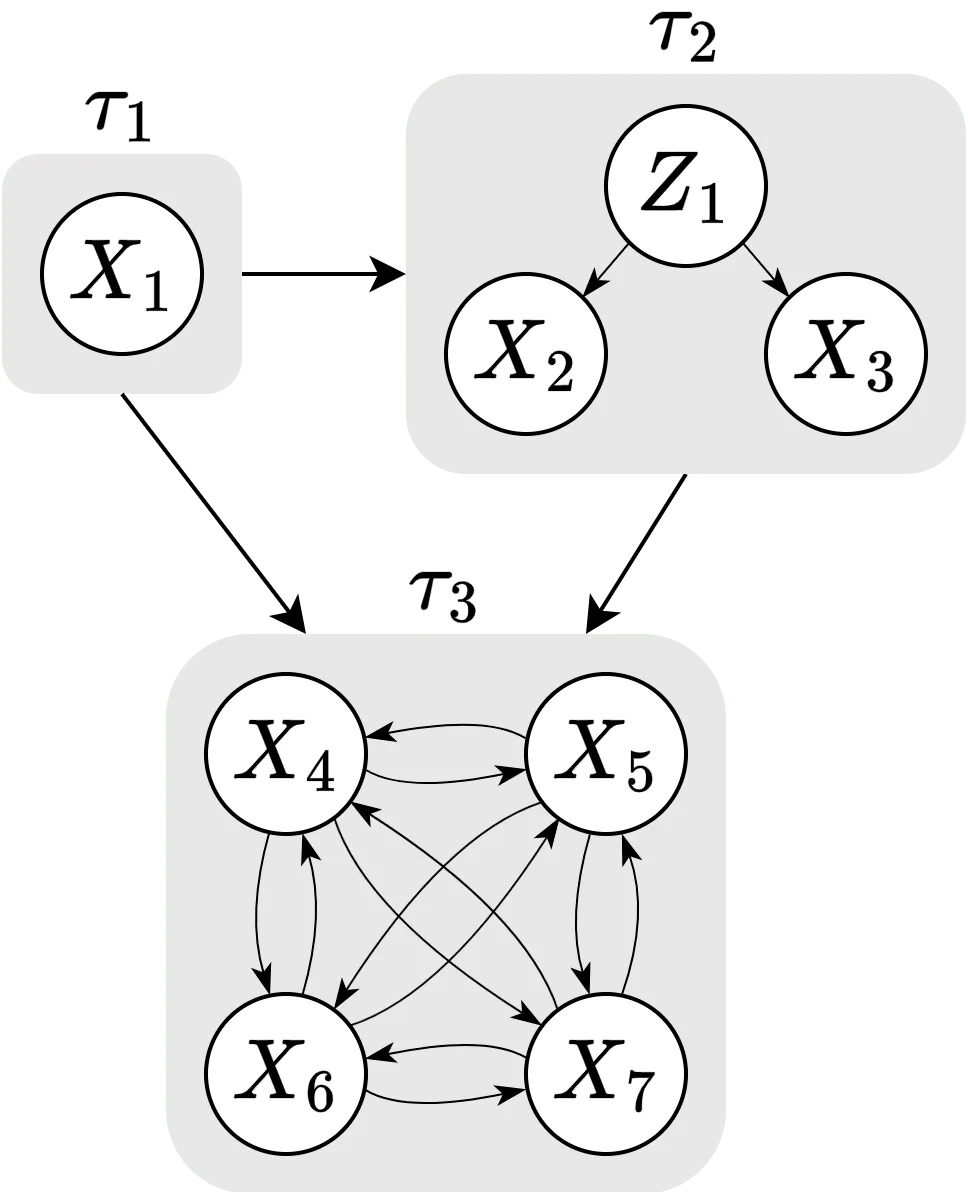
Schematic overview of the causal chain graph used in this vignette.
Marginal Shapley values
Causal Shapley values are equivalent to marginal Shapley values when all features are combined into a single component and all dependencies are induced by confounding. Then , and in Equation () simplifies to , as specified in Lundberg and Lee (2017).
The Monte Carlo samples for the marginals are generated by sampling
from the training data, except for the gaussian approach
where we use the marginals of the estimated multivariate Gaussian
distribution. This means that for all other approaches, this is the same
as using the independence approach in the conditional
Shapley value explanation framework.
Symmetric conditional Shapley values
Causal Shapley values are equivalent to symmetric conditional Shapley values when all features are combined into a single component and all dependencies are induced by mutual interaction. Then , and in Equation () simplifies to , as specified in Aas, Jullum, and Løland (2021). Symmetric means that we consider all coalitions.
Code example
Overview
We demonstrate the frameworks on the bike sharing dataset
from the UCI Machine Learning Repository. We let the features be the
number of days since January 2011 (trend), two cyclical
variables representing the season (cosyear,
sinyear), temperature (temp), feeling
temperature (atemp), wind speed (windspeed),
and humidity (hum). The first three features are considered
to be a potential cause of the four weather-related features. The bike
rental is strongly seasonal and shows an upward trend, as illustrated in
the figure below. The bike data is split randomly into a training (80%)
and test/explicand (20%) set. We train an XGBoost model for
100 rounds with default hyperparameters to act as the model we want to
explain.
In the table below, we highlight the Shapley value explanation
frameworks introduced above and how to access them by changing the
arguments asymmetric, ordering, and
confounding in shapr::explain(). Note that
symmetric conditional Shapley values are the default version; i.e., by
default asymmetric = FALSE, ordering = NULL,
confounding = NULL.
| Framework | Sampling | Approaches | asymmetric |
ordering |
confounding |
|---|---|---|---|---|---|
| Sym. Conditional | All | FALSE |
NULL |
NULL |
|
| Asym. Conditional | All | TRUE |
list(...) |
NULL |
|
| Sym. Causal | All MC-based | FALSE |
list(...) |
c(...) |
|
| Asym. Causal | All MC-based | TRUE |
list(...) |
c(...) |
|
| Sym. Marginal |
indep., gaussian
|
FALSE |
NULL |
TRUE |
Code setup
First, we load the needed libraries, set up the training/explicand
data, plot the data, and train an xgboost model.
library(ggplot2)
library(xgboost)
library(data.table)
library(shapr)
# Additional packages which are only used for plotting in this vignette.
# These are not listed as dependencies in shapr
library(GGally)
library(ggpubr)
library(gridExtra)
# Ensure that shapr's functions are prioritized; otherwise, we need to use the `shapr::`
# prefix when calling explain(). The `conflicted` package is imported by `tidymodels`.
conflicted::conflicts_prefer(shapr::explain, shapr::prepare_data)
# Set up the data
# Can also download the data set from the source https://archive.ics.uci.edu/dataset/275/bike+sharing+dataset
# temp <- tempfile()
# download.file("https://archive.ics.uci.edu/static/public/275/bike+sharing+dataset.zip", temp)
# bike <- read.csv(unz(temp, "day.csv"))
# unlink(temp)
bike <- read.csv("../inst/extdata/day.csv")
# Difference in days, which takes DST into account
bike$trend <- as.numeric(difftime(bike$dteday, bike$dteday[1], units = "days"))
bike$cosyear <- cospi(bike$trend / 365 * 2)
bike$sinyear <- sinpi(bike$trend / 365 * 2)
# Unnormalize variables (see data set information in link above)
bike$temp <- bike$temp * (39 - (-8)) + (-8)
bike$atemp <- bike$atemp * (50 - (-16)) + (-16)
bike$windspeed <- 67 * bike$windspeed
bike$hum <- 100 * bike$hum
# Plot the data
ggplot(bike, aes(x = trend, y = cnt, color = temp)) +
geom_point(size = 0.75) +
scale_color_gradient(low = "blue", high = "red") +
labs(colour = "temp") +
xlab("Days since 1 January 2011") +
ylab("Number of bikes rented") +
theme_minimal() +
theme(legend.position = "right", legend.title = element_text(size = 10))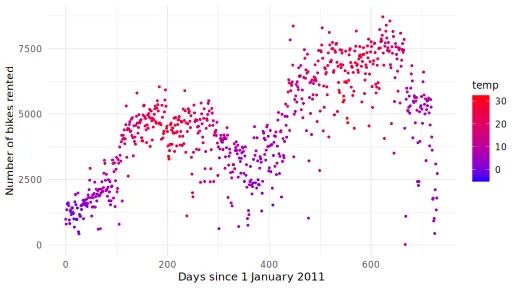
# Define the features and the response variable
x_var <- c("trend", "cosyear", "sinyear", "temp", "atemp", "windspeed", "hum")
y_var <- "cnt"
# NOTE: To avoid RNG reproducibility issues across different systems, we
# load the training-test split from a file. 80% training and 20% test
train_index <- readRDS("../inst/extdata/train_index.rds")
# Training data
x_train <- as.matrix(bike[train_index, x_var])
y_train_nc <- as.matrix(bike[train_index, y_var]) # not centered
y_train <- y_train_nc - mean(y_train_nc)
# Plot pairs plot
GGally::ggpairs(x_train)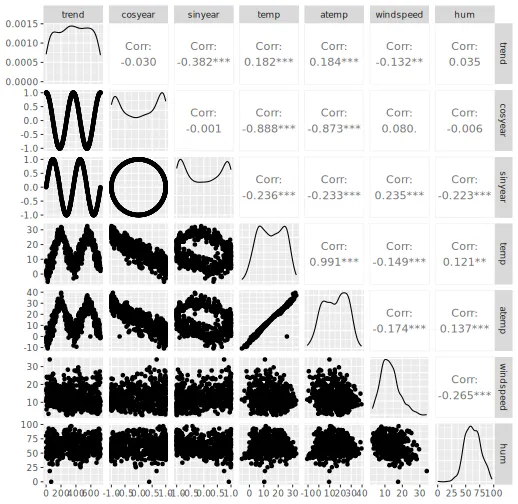
# Test/explicand data
x_explain <- as.matrix(bike[-train_index, x_var])
y_explain_nc <- as.matrix(bike[-train_index, y_var]) # not centered
y_explain <- y_explain_nc - mean(y_train_nc)
# Get 6 explicands to plot the Shapley values for, with a wide spread in their predicted outcomes
n_index_x_explain <- 6
index_x_explain <- order(y_explain)[seq(1, length(y_explain), length.out = n_index_x_explain)]
y_explain[index_x_explain]
#> [1] -3900.03 -1872.03 -377.03 411.97 1690.97 3889.97
# Fit an XGBoost model to the training data
model <- xgboost::xgboost(
x = x_train,
y = y_train,
nround = 100,
verbosity = 0
)
# Save the phi0
phi0 <- mean(y_train)
# Look at the root mean squared error
sqrt(mean((predict(model, x_explain) - y_explain)^2))
#> [1] 768.96
ggplot(
data.table("response" = y_explain[, 1], "predicted_response" = predict(model, x_explain)),
aes(response, predicted_response)
) +
geom_point()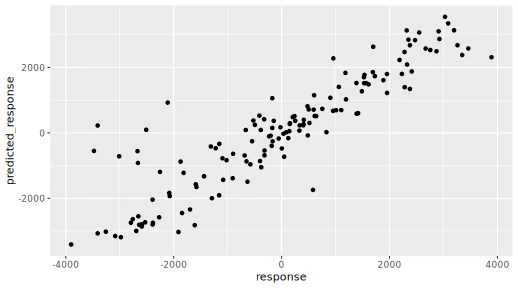
We are going to use the causal_ordering and
confounding illustrated in the figures above. For
causal_ordering, we can either provide the index of feature
or the feature names. Thus, the following two versions of
causal_ordering will produce equivalent results.
Furthermore, we assume that we have confounding for the second component
(i.e., the season has an effect on the weather) and no confounding for
the third component (i.e., we do not know how to model the intricate
relations between the weather features).
causal_ordering <- list(1, c(2, 3), c(4:7))
causal_ordering <- list("trend", c("cosyear", "sinyear"), c("temp", "atemp", "windspeed", "hum"))
confounding <- c(FALSE, TRUE, FALSE)To make the rest of the vignette easier to follow, we create some helper functions that plot and summarize the results of the explanation methods. This code block is optional to understand and can be skipped.
# Extract the MSEv criterion scores and elapsed times
print_MSEv_scores_and_time <- function(explanation_list) {
res <- as.data.frame(t(sapply(
explanation_list,
function(explanation) {
round(c(
explanation$MSEv$MSEv$MSEv,
explanation$MSEv$MSEv$MSEv_sd,
explanation$timing$summary$total_time_secs
), 2)
}
)))
colnames(res) <- c("MSEv", "MSEv_sd", "Time (secs)")
return(res)
}
# Print the full time information
print_time <- function(explanation_list) {
t(sapply(explanation_list, function(explanation) explanation$timing$summary$total_time_secs))
}
# Make beeswarm plots
plot_beeswarms <- function(explanation_list, title = "", ...) {
# Make the beeswarm plots
grobs <- lapply(seq(length(explanation_list)), function(explanation_idx) {
gg <- suppressMessages(
plot(explanation_list[[explanation_idx]], plot_type = "beeswarm", print_ggplot = FALSE, ...)
) +
ggplot2::ggtitle(tools::toTitleCase(gsub("_", " ", names(explanation_list)[[explanation_idx]])))
# Flip the order such that the features come in the right order
gg <- gg +
ggplot2::scale_x_discrete(limits = rev(levels(gg$data$variable)[levels(gg$data$variable) != "none"]))
})
# Get the limits
ylim <- sapply(grobs, function(grob) ggplot2::ggplot_build(grob)$layout$panel_scales_y[[1]]$range$range)
ylim <- c(min(ylim), max(ylim))
# Update the limits
grobs <- suppressMessages(lapply(grobs, function(grob) grob + ggplot2::coord_flip(ylim = ylim)))
# Make the combined plot
gridExtra::grid.arrange(
grobs = grobs, ncol = 1,
top = grid::textGrob(title, gp = grid::gpar(fontsize = 18, font = 8))
)
}Symmetric conditional Shapley values (default)
We start by demonstrating how to compute symmetric conditional
Shapley values. This is the default version in shapr and
there is no need to specify the arguments below. However, we have
specified them for the sake of clarity. We use the
gaussian, ctree, and
regression_separate(xgboost with default
hyperparameters) approaches, but any other approach can also be
used.
# list to store the results
explanation_sym_con <- list()
explanation_sym_con[["gaussian"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
approach = "gaussian",
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
asymmetric = FALSE, # Default value (TRUE will give the same since `causal_ordering = NULL`)
causal_ordering = NULL, # Default value
confounding = NULL # Default value
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:57:35
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 128`, and is therefore set to
#> `2^n_features = 128`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: gaussian
#>
#> • Procedure: Iterative
#>
#> • Number of Monte Carlo integration samples: 1000
#>
#> • Number of feature-wise Shapley values: 7
#>
#> • Number of observations to explain: 144
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16486325de97.rds'
#>
#>
#>
#> ── Iterative computation started ──
#>
#>
#>
#> ── Iteration 1 ───────────────────────────────────────
#>
#> ℹ Using 14 of 128 coalitions, 14 new.
#>
#>
#>
#> ── Iteration 2 ───────────────────────────────────────
#>
#> ℹ Using 26 of 128 coalitions, 12 new.
#>
#>
#>
#> ── Iteration 3 ───────────────────────────────────────
#>
#> ℹ Using 46 of 128 coalitions, 20 new.
#>
#>
#>
#> ── Iteration 4 ───────────────────────────────────────
#>
#> ℹ Using 68 of 128 coalitions, 22 new.
explanation_sym_con[["ctree"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
approach = "ctree",
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
asymmetric = FALSE, # Default value (TRUE will give the same since `causal_ordering = NULL`)
causal_ordering = NULL, # Default value
confounding = NULL # Default value
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:57:47
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 128`, and is therefore set to
#> `2^n_features = 128`.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: ctree
#> • Procedure: Iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16484e66376.rds'
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ───────────────────────────────────────
#> ℹ Using 14 of 128 coalitions, 14 new.
#>
#> ── Iteration 2 ───────────────────────────────────────
#> ℹ Using 26 of 128 coalitions, 12 new.
#>
#> ── Iteration 3 ───────────────────────────────────────
#> ℹ Using 46 of 128 coalitions, 20 new.
#>
#> ── Iteration 4 ───────────────────────────────────────
#> ℹ Using 70 of 128 coalitions, 24 new.
#>
#> ── Iteration 5 ───────────────────────────────────────
#> ℹ Using 94 of 128 coalitions, 24 new.
#>
#> ── Iteration 6 ───────────────────────────────────────
#> ℹ Using 108 of 128 coalitions, 14 new.
explanation_sym_con[["xgboost"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
approach = "regression_separate",
regression.model = parsnip::boost_tree(engine = "xgboost", mode = "regression"),
asymmetric = FALSE, # Default value (TRUE will give the same as `causal_ordering = NULL`)
causal_ordering = NULL, # Default value
confounding = NULL # Default value
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:58:56
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 128`, and is therefore set to
#> `2^n_features = 128`.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Regression
#> • Approach: regression_separate
#> • Procedure: Iterative
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164851bcd272.rds'
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ───────────────────────────────────────
#> ℹ Using 14 of 128 coalitions, 14 new.
#>
#> ── Iteration 2 ───────────────────────────────────────
#> ℹ Using 26 of 128 coalitions, 12 new.
#>
#> ── Iteration 3 ───────────────────────────────────────
#> ℹ Using 46 of 128 coalitions, 20 new.
#>
#> ── Iteration 4 ───────────────────────────────────────
#> ℹ Using 70 of 128 coalitions, 24 new.
#>
#> ── Iteration 5 ───────────────────────────────────────
#> ℹ Using 94 of 128 coalitions, 24 new.
#>
#> ── Iteration 6 ───────────────────────────────────────
#> ℹ Using 110 of 128 coalitions, 16 new.
#>
#> ── Iteration 7 ───────────────────────────────────────
#> ℹ Using 114 of 128 coalitions, 4 new.We can then look at the
evaluation scores to compare the approaches. All approaches are
comparable, but xgboost is clearly the fastest
approach.
print_MSEv_scores_and_time(explanation_sym_con)
#> MSEv MSEv_sd Time (secs)
#> gaussian 1101051 71271 11.79
#> ctree 1060934 63108 68.60
#> xgboost 1090931 58988 18.73We can then plot the Shapley values for the six explicands chosen above.
plot_SV_several_approaches(explanation_sym_con, index_x_explain, print_ggplot = FALSE) +
theme(legend.position = "bottom")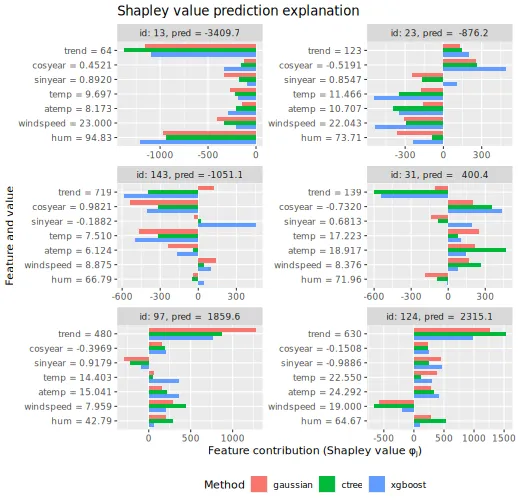
We can also make beeswarm plots of the Shapley values to look at the
structure of the Shapley values for all explicands. The figures are
quite similar, but with minor differences. E.g., the
gaussian approach produces almost no Shapley values around
for the trend feature.
plot_beeswarms(explanation_sym_con, title = "Symmetric conditional Shapley values")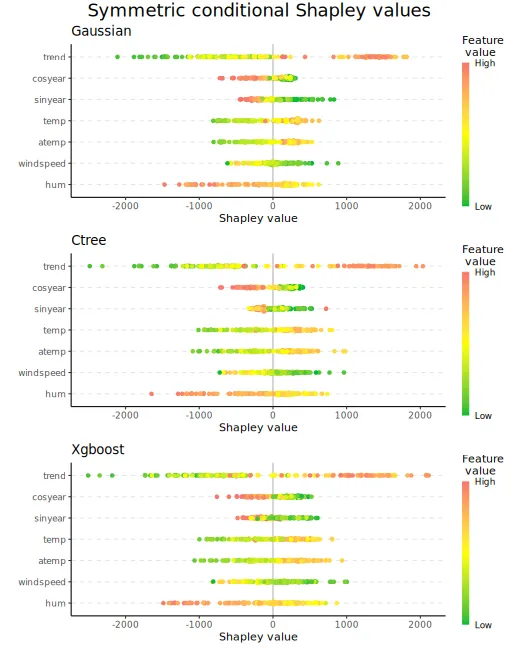
Asymmetric conditional Shapley values
Then we look at the asymmetric conditional Shapley values. To obtain
these types of Shapley values, we have to specify that
asymmetric = TRUE and a causal_ordering. We
use causal_ordering = list(1, c(2, 3), c(4:7)).
explanation_asym_con <- list()
explanation_asym_con[["gaussian"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "gaussian",
asymmetric = TRUE,
causal_ordering = causal_ordering,
confounding = NULL # Default value
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:59:17
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than the
#> number of coalitions respecting the causal ordering
#> (20), and is therefore set to 20.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: gaussian
#>
#> • Procedure: Iterative
#>
#> • Number of Monte Carlo integration samples: 1000
#>
#> • Number of feature-wise Shapley values: 7
#>
#> • Number of observations to explain: 144
#>
#> • Number of asymmetric coalitions: 20
#>
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164866a43c71.rds'
#>
#>
#>
#> ── Iterative computation started ──
#>
#>
#>
#> ── Iteration 1 ───────────────────────────────────────
#>
#> ℹ Using 13 of 20 coalitions, 13 new.
#>
#>
#>
#> ── Iteration 2 ───────────────────────────────────────
#>
#> ℹ Using 20 of 20 coalitions, 7 new.
explanation_asym_con[["gaussian_non_iterative"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "gaussian",
asymmetric = TRUE,
causal_ordering = causal_ordering,
confounding = NULL, # Default value
iterative = FALSE
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:59:21
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than the
#> number of coalitions respecting the causal ordering
#> (20), and is therefore set to 20.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Number of asymmetric coalitions: 20
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16487c406765.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 20 of 20 coalitions.
explanation_asym_con[["ctree"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "ctree",
asymmetric = TRUE,
causal_ordering = causal_ordering,
confounding = NULL # Default value
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:59:23
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than the
#> number of coalitions respecting the causal ordering
#> (20), and is therefore set to 20.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: ctree
#> • Procedure: Iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Number of asymmetric coalitions: 20
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16481bcde567.rds'
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ───────────────────────────────────────
#> ℹ Using 13 of 20 coalitions, 13 new.
#>
#> ── Iteration 2 ───────────────────────────────────────
#> ℹ Using 20 of 20 coalitions, 7 new.
explanation_asym_con[["xgboost"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
approach = "regression_separate",
regression.model = parsnip::boost_tree(engine = "xgboost", mode = "regression"),
asymmetric = TRUE,
causal_ordering = causal_ordering,
confounding = NULL # Default value
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:59:35
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than the
#> number of coalitions respecting the causal ordering
#> (20), and is therefore set to 20.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Regression
#> • Approach: regression_separate
#> • Procedure: Iterative
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Number of asymmetric coalitions: 20
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164865579755.rds'
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ───────────────────────────────────────
#> ℹ Using 13 of 20 coalitions, 13 new.
#>
#> ── Iteration 2 ───────────────────────────────────────
#> ℹ Using 20 of 20 coalitions, 7 new.The asymmetric conditional Shapley value framework is faster as we only consider coalitions (including the empty and grand coalitions) instead of all coalitions (see code below).
print_MSEv_scores_and_time(explanation_asym_con)
#> MSEv MSEv_sd Time (secs)
#> gaussian 339662 38586 4.75
#> gaussian_non_iterative 339521 38588 1.68
#> ctree 282686 32264 12.26
#> xgboost 321763 39778 3.85
# Look at the number of coalitions considered. Decreased from 128 to 20.
explanation_sym_con$gaussian$internal$parameters$max_n_coalitions
#> [1] 128
explanation_asym_con$gaussian$internal$parameters$max_n_coalitions
#> [1] 20
# Here we can see the 20 coalitions that respect the causal ordering
explanation_asym_con$gaussian$internal$objects$dt_valid_causal_coalitions[["coalitions"]]
#> [[1]]
#> integer(0)
#>
#> [[2]]
#> [1] 1
#>
#> [[3]]
#> [1] 1 2
#>
#> [[4]]
#> [1] 1 3
#>
#> [[5]]
#> [1] 1 2 3
#>
#> [[6]]
#> [1] 1 2 3 4
#>
#> [[7]]
#> [1] 1 2 3 5
#>
#> [[8]]
#> [1] 1 2 3 6
#>
#> [[9]]
#> [1] 1 2 3 7
#>
#> [[10]]
#> [1] 1 2 3 4 5
#>
#> [[11]]
#> [1] 1 2 3 4 6
#>
#> [[12]]
#> [1] 1 2 3 4 7
#>
#> [[13]]
#> [1] 1 2 3 5 6
#>
#> [[14]]
#> [1] 1 2 3 5 7
#>
#> [[15]]
#> [1] 1 2 3 6 7
#>
#> [[16]]
#> [1] 1 2 3 4 5 6
#>
#> [[17]]
#> [1] 1 2 3 4 5 7
#>
#> [[18]]
#> [1] 1 2 3 4 6 7
#>
#> [[19]]
#> [1] 1 2 3 5 6 7
#>
#> [[20]]
#> [1] 1 2 3 4 5 6 7We can then look at the beeswarm plots of the asymmetric conditional
Shapley values. The ctree and xgboost
approaches produce similar figures, while the gaussian
approach shrinks and groups the Shapley values for the
trend feature, and it produces more negative values for the
cosyear feature.
When going from symmetric to asymmetric Shapley values, we see that
many of the features’ Shapley values have now shrunk closer to zero,
especially temp and atemp.
plot_beeswarms(explanation_asym_con, title = "Asymmetric conditional Shapley values")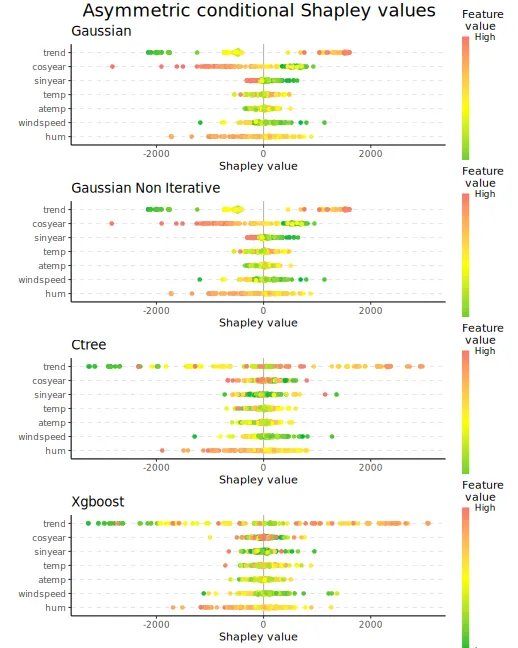
We can also compare the obtained symmetric and asymmetric conditional
Shapley values for the 6 explicands. We often see that the asymmetric
version gives larger Shapley values to the distal/root causes, i.e.,
trend and cosyear, than the symmetric version.
This is in line with Section 3.2 in Frye, Rowat,
and Feige (2020).
# Order the symmetric and asymmetric conditional explanations into a joint list
explanation_sym_con_tmp <- copy(explanation_sym_con)
names(explanation_sym_con_tmp) <- paste0(names(explanation_sym_con_tmp), "_sym")
explanation_asym_con_tmp <- copy(explanation_asym_con)
names(explanation_asym_con_tmp) <- paste0(names(explanation_asym_con_tmp), "_asym")
explanation_asym_sym_con <- c(explanation_sym_con_tmp, explanation_asym_con_tmp)[c(1, 4, 2, 5, 3, 6)]
plot_SV_several_approaches(explanation_asym_sym_con, index_x_explain, brewer_palette = "Paired", print_ggplot = FALSE) +
theme(legend.position = "bottom")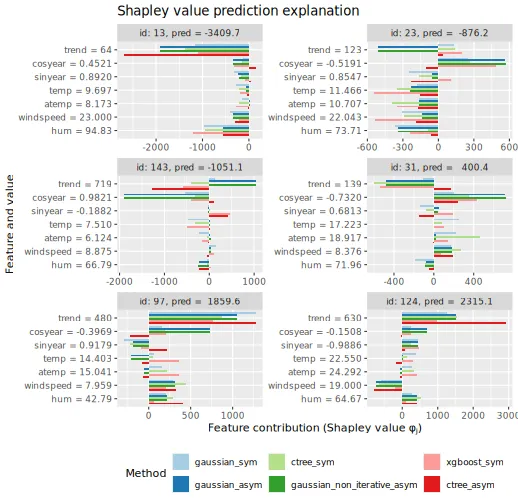
Symmetric marginal Shapley values
For marginal Shapley values, we can only consider the symmetric
version as we must set causal_ordering = list(1:7) (or
NULL) and confounding = TRUE. Setting
asymmetric = TRUE will have no effect, as the causal
ordering consists of only a single component containing all features,
i.e., all coalitions respect the causal ordering. As stated above,
shapr generates the marginal Monte Carlo samples from the
Gaussian marginals if approach = "gaussian", while for all
other Monte Carlo approaches the marginals are estimated from the
training data, i.e., assuming feature independence. Thus, it does not
matter if we set approach = "independence" or any other of
the Monte Carlo-based approaches. We use
approach = "independence" for clarity. Furthermore, we also
obtain marginal Shapley values by using the conditional Shapley value
framework with the independence approach. However, note
that there will be a minuscule difference in the produced Shapley values
due to different sampling setups/orders.
explanation_sym_marg <- list()
# Here we sample from the estimated Gaussian marginals
explanation_sym_marg[["gaussian"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "gaussian",
asymmetric = FALSE,
causal_ordering = list(1:7),
confounding = TRUE
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:59:42
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 128`, and is therefore set to
#> `2^n_features = 128`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: gaussian
#>
#> • Procedure: Iterative
#>
#> • Number of Monte Carlo integration samples: 1000
#>
#> • Number of feature-wise Shapley values: 7
#>
#> • Number of observations to explain: 144
#>
#> • Causal ordering: {trend, cosyear, sinyear, temp,
#> atemp, windspeed, hum}
#>
#> • Components with confounding: {trend, cosyear,
#> sinyear, temp, atemp, windspeed, hum}
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164838d38a04.rds'
#>
#>
#>
#> ── Iterative computation started ──
#>
#>
#>
#> ── Iteration 1 ───────────────────────────────────────
#>
#> ℹ Using 14 of 128 coalitions, 14 new.
#>
#>
#>
#> ── Iteration 2 ───────────────────────────────────────
#>
#> ℹ Using 26 of 128 coalitions, 12 new.
#>
#>
#>
#> ── Iteration 3 ───────────────────────────────────────
#>
#> ℹ Using 46 of 128 coalitions, 20 new.
# Here we sample from the marginals of the training data
explanation_sym_marg[["independence_marg"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "independence",
asymmetric = FALSE,
causal_ordering = list(1:7),
confounding = TRUE
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 12:59:52
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 128`, and is therefore set to
#> `2^n_features = 128`.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: independence
#> • Procedure: Iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Causal ordering: {trend, cosyear, sinyear, temp,
#> atemp, windspeed, hum}
#> • Components with confounding: {trend, cosyear,
#> sinyear, temp, atemp, windspeed, hum}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164838bdee89.rds'
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ───────────────────────────────────────
#> ℹ Using 14 of 128 coalitions, 14 new.
#>
#> ── Iteration 2 ───────────────────────────────────────
#> ℹ Using 26 of 128 coalitions, 12 new.
#>
#> ── Iteration 3 ───────────────────────────────────────
#> ℹ Using 46 of 128 coalitions, 20 new.
# Here we use the conditional Shapley value framework with the `independence` approach
explanation_sym_marg[["independence_con"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "independence"
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:00:00
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 128`, and is therefore set to
#> `2^n_features = 128`.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: independence
#> • Procedure: Iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16482c3c2bd6.rds'
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ───────────────────────────────────────
#> ℹ Using 14 of 128 coalitions, 14 new.
#>
#> ── Iteration 2 ───────────────────────────────────────
#> ℹ Using 26 of 128 coalitions, 12 new.
#>
#> ── Iteration 3 ───────────────────────────────────────
#> ℹ Using 46 of 128 coalitions, 20 new.We can look at the beeswarm plots
print_MSEv_scores_and_time(explanation_sym_marg)
#> MSEv MSEv_sd Time (secs)
#> gaussian 1396525 109151 9.95
#> independence_marg 1390794 107977 7.95
#> independence_con 1390287 107899 10.67
plot_beeswarms(explanation_sym_marg, title = "Symmetric marginal Shapley values")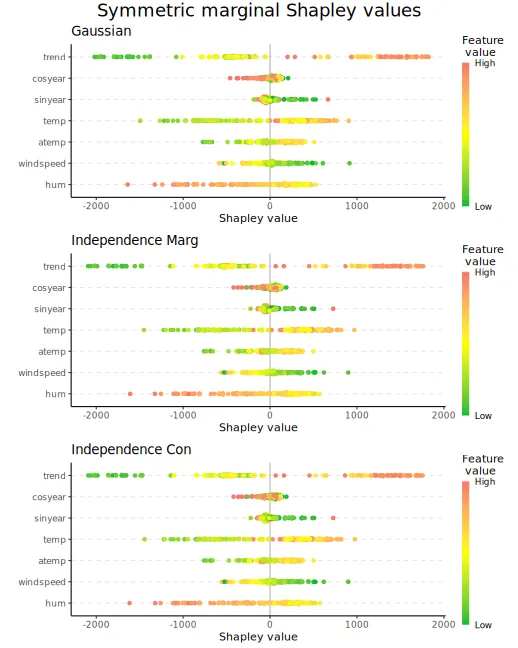
Causal Shapley values
To compute (symmetric/asymmetric) causal Shapley values, we have to
provide the causal_ordering and confounding
objects. We set them to be
causal_ordering = list(1, 2:3, 4:7) and
confounding = c(FALSE, TRUE, FALSE), as explained
above.
The causal framework takes longer than the other frameworks, as
generating the Monte Carlo samples often consists of a chain of sampling
steps. For example, for
,
we must generate
.
However, we cannot do this directly due to the
causal_ordering and confounding specified
above. To generate the Monte Carlo samples, we have to follow a chain of
sampling steps. More precisely, we first need to generate
from the marginal, then
,
and finally
.
The latter two steps are done by using the provided
approach to model the conditional distributions. The
internal$objects$S_causal_steps_strings object contains the
sampling steps needed for the different feature combinations/coalitions
.
For causal Shapley values, only the Monte Carlo-based approaches are applicable.
Symmetric
explanation_sym_cau <- list()
explanation_sym_cau[["gaussian"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "gaussian",
asymmetric = FALSE,
causal_ordering = list(1, 2:3, 4:7),
confounding = c(FALSE, TRUE, FALSE),
iterative = FALSE, # Set to FALSE to get a single iteration to illustrate sampling steps below
exact = TRUE
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:00:12
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 128`, and is therefore set to
#> `2^n_features = 128`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: gaussian
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 1000
#>
#> • Number of feature-wise Shapley values: 7
#>
#> • Number of observations to explain: 144
#>
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#>
#> • Components with confounding: {cosyear, sinyear}
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_1648e02fb73.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 128 of 128 coalitions.
# Look at the sampling steps for the third coalition (S = {2})
explanation_sym_cau$gaussian$internal$iter_list[[1]]$S_causal_steps_strings$id_coalition_3
#> [1] "1|" "3|1" "4,5,6,7|1,2,3"
# Use the copula approach
explanation_sym_cau[["copula"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "copula",
asymmetric = FALSE,
causal_ordering = list(1, 2:3, 4:7),
confounding = c(FALSE, TRUE, FALSE)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:00:48
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_features = 128`, and is therefore set to
#> `2^n_features = 128`.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: copula
#> • Procedure: Iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#> • Components with confounding: {cosyear, sinyear}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16485ccc1bb0.rds'
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ───────────────────────────────────────
#> ℹ Using 14 of 128 coalitions, 14 new.
#>
#> ── Iteration 2 ───────────────────────────────────────
#> ℹ Using 26 of 128 coalitions, 12 new.
#>
#> ── Iteration 3 ───────────────────────────────────────
#> ℹ Using 46 of 128 coalitions, 20 new.
print_MSEv_scores_and_time(explanation_sym_cau)
#> MSEv MSEv_sd Time (secs)
#> gaussian 1147280 83432 35.70
#> copula 1166326 86145 26.95
plot_beeswarms(explanation_sym_cau, title = "Symmetric causal Shapley values")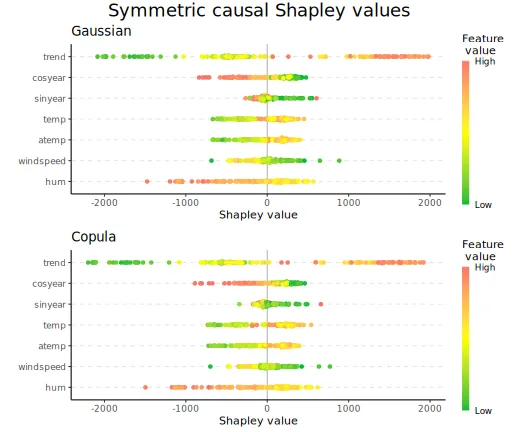
Asymmetric
We now turn to asymmetric causal Shapley values. That is, we only use the coalitions that respect the causal ordering. Thus, the computations are faster as the number of coalitions is reduced.
explanation_asym_cau <- list()
explanation_asym_cau[["gaussian"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "gaussian",
asymmetric = TRUE,
causal_ordering = list(1, 2:3, 4:7),
confounding = c(FALSE, TRUE, FALSE)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:01:15
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than the
#> number of coalitions respecting the causal ordering
#> (20), and is therefore set to 20.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: gaussian
#>
#> • Procedure: Iterative
#>
#> • Number of Monte Carlo integration samples: 1000
#>
#> • Number of feature-wise Shapley values: 7
#>
#> • Number of observations to explain: 144
#>
#> • Number of asymmetric coalitions: 20
#>
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#>
#> • Components with confounding: {cosyear, sinyear}
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_1648212ff635.rds'
#>
#>
#>
#> ── Iterative computation started ──
#>
#>
#>
#> ── Iteration 1 ───────────────────────────────────────
#>
#> ℹ Using 13 of 20 coalitions, 13 new.
#>
#>
#>
#> ── Iteration 2 ───────────────────────────────────────
#>
#> ℹ Using 20 of 20 coalitions, 7 new.
# Use the copula approach
explanation_asym_cau[["copula"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "copula",
asymmetric = TRUE,
causal_ordering = list(1, 2:3, 4:7),
confounding = c(FALSE, TRUE, FALSE)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:01:21
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than the
#> number of coalitions respecting the causal ordering
#> (20), and is therefore set to 20.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: copula
#> • Procedure: Iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Number of asymmetric coalitions: 20
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#> • Components with confounding: {cosyear, sinyear}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16486793ff26.rds'
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ───────────────────────────────────────
#> ℹ Using 13 of 20 coalitions, 13 new.
#>
#> ── Iteration 2 ───────────────────────────────────────
#> ℹ Using 20 of 20 coalitions, 7 new.
# Use the vaeac approach
explanation_asym_cau[["vaeac"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
phi0 = phi0,
seed = 1,
n_MC_samples = 1000,
approach = "vaeac",
vaeac.epochs = 20,
asymmetric = TRUE,
causal_ordering = list(1, 2:3, 4:7),
confounding = c(FALSE, TRUE, FALSE)
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:01:26
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than the
#> number of coalitions respecting the causal ordering
#> (20), and is therefore set to 20.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: vaeac
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Number of asymmetric coalitions: 20
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#> • Components with confounding: {cosyear, sinyear}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16483556ca80.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 20 of 20 coalitions.We can look at the elapsed time. See the implementation details for an explanation.
print_time(explanation_asym_cau)
#> gaussian copula vaeac
#> [1,] 5.4413 4.8707 332.07We can then plot the beeswarm plots.
# Plot the beeswarm plots
plot_beeswarms(explanation_asym_cau, title = "Asymmetric causal Shapley values")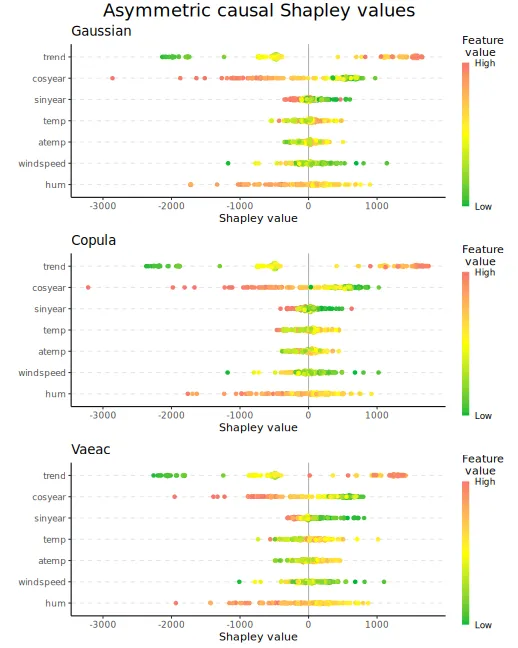
# Plot the Shapley values
plot_SV_several_approaches(explanation_asym_cau, index_x_explain, print_ggplot = FALSE) +
theme(legend.position = "bottom")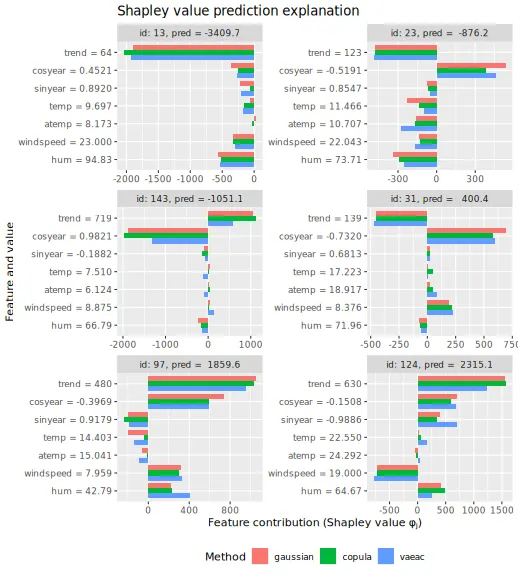
We can also use the other Monte Carlo-based approaches
(independence and empirical), too.
Comparing the frameworks
Here we plot the obtained Shapley values for the six explicands when
using the gaussian approach in the different Shapley value
explanation frameworks, and we see that the different frameworks provide
different explanations. The largest difference is between the symmetric
and asymmetric versions. To summarize, asymmetric conditional/causal
Shapley values focus on the root cause, marginal Shapley values on the
more direct effect, and symmetric conditional/causal Shapley values
consider both for a more natural explanation.
explanation_gaussian <- list(
symmetric_marginal = explanation_sym_marg$gaussian,
symmetric_conditional = explanation_sym_con$gaussian,
symmetric_causal = explanation_sym_cau$gaussian,
asymmetric_conditional = explanation_asym_con$gaussian,
asymmetric_causal = explanation_asym_cau$gaussian
)
plot_SV_several_approaches(explanation_gaussian, index_x_explain, print_ggplot = FALSE) +
theme(legend.position = "bottom") +
guides(fill = guide_legend(nrow = 2)) +
ggtitle("Shapley value prediction explanation (approach = 'gaussian')") +
guides(color = guide_legend(title = "Framework"))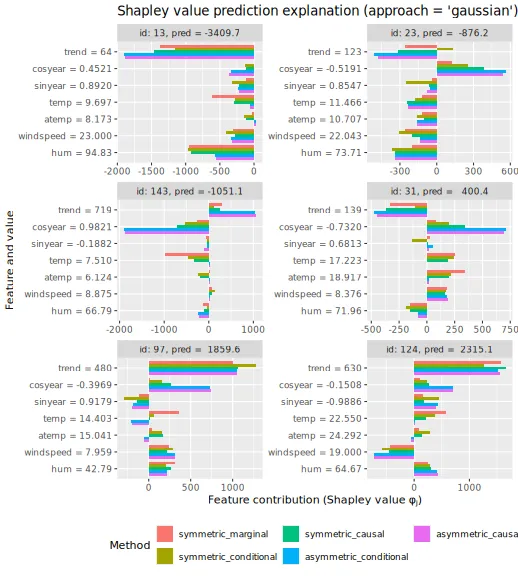
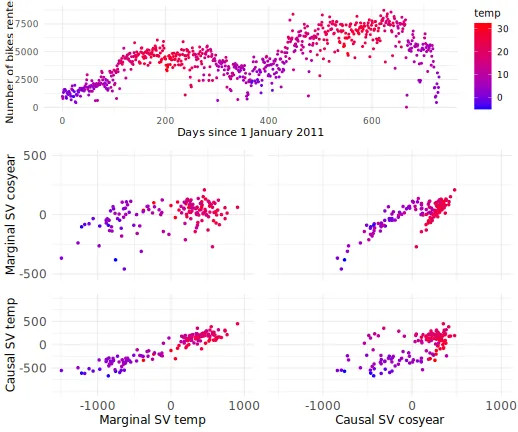
Scatter plots: marginal vs. causal Shapley values
In this section, we produce scatter plots comparing the symmetric
marginal and symmetric causal Shapley values for the temperature feature
temp and the seasonal feature cosyear for all
explicands. The plots show that the marginal Shapley values almost
purely explain the predictions based on temperature, while the causal
Shapley values also give credit to season. We can change the features
and frameworks in the code below, but we chose these values to replicate
Figure 3 in Heskes et al. (2020).
# The color of the points
color <- "temp"
# The features we want to compare
feature_1 <- "cosyear"
feature_2 <- "temp"
# The Shapley value frameworks we want to compare
sv_framework_1 <- explanation_sym_marg[["gaussian"]]
sv_framework_1_str <- "Marginal SV"
sv_framework_2 <- explanation_sym_cau[["gaussian"]]
sv_framework_2_str <- "Causal SV"
# Set up the data.frame we are going to plot
sv_correlation_df <- data.frame(
color = x_explain[, color],
sv_framework_1_feature_1 = sv_framework_1$shapley_values_est[[feature_1]],
sv_framework_2_feature_1 = sv_framework_2$shapley_values_est[[feature_1]],
sv_framework_1_feature_2 = sv_framework_1$shapley_values_est[[feature_2]],
sv_framework_2_feature_2 = sv_framework_2$shapley_values_est[[feature_2]]
)
# Make the plots
scatterplot_topleft <-
ggplot(
sv_correlation_df,
aes(x = sv_framework_1_feature_2, y = sv_framework_1_feature_1, color = color)
) +
geom_point(size = 1) +
xlab(paste(sv_framework_1_str, feature_2)) +
ylab(paste(sv_framework_1_str, feature_1)) +
scale_x_continuous(limits = c(-1500, 1000), breaks = c(-1000, 0, 1000)) +
scale_y_continuous(limits = c(-500, 500), breaks = c(-500, 0, 500)) +
scale_color_gradient(low = "blue", high = "red") +
theme_minimal() +
theme(
text = element_text(size = 12),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 12),
axis.ticks.x = element_blank(),
axis.title.x = element_blank()
)
scatterplot_topright <-
ggplot(
sv_correlation_df,
aes(x = sv_framework_2_feature_1, y = sv_framework_1_feature_1, color = color)
) +
geom_point(size = 1) +
scale_color_gradient(low = "blue", high = "red") +
xlab(paste(sv_framework_2_str, feature_1)) +
ylab(paste(sv_framework_1_str, feature_1)) +
scale_x_continuous(limits = c(-1500, 1000), breaks = c(-1000, 0, 1000)) +
scale_y_continuous(limits = c(-500, 500), breaks = c(-500, 0, 500)) +
theme_minimal() +
theme(
text = element_text(size = 12),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank()
)
scatterplot_bottomleft <-
ggplot(
sv_correlation_df,
aes(x = sv_framework_1_feature_2, y = sv_framework_2_feature_2, color = color)
) +
geom_point(size = 1) +
scale_color_gradient(low = "blue", high = "red") +
xlab(paste(sv_framework_1_str, feature_2)) +
ylab(paste(sv_framework_2_str, feature_2)) +
scale_x_continuous(limits = c(-1500, 1000), breaks = c(-1000, 0, 1000)) +
scale_y_continuous(limits = c(-1000, 1000), breaks = c(-500, 0, 500)) +
theme_minimal() +
theme(
text = element_text(size = 12),
axis.text.x = element_text(size = 12),
axis.text.y = element_text(size = 12)
)
scatterplot_bottomright <-
ggplot(
sv_correlation_df,
aes(x = sv_framework_2_feature_1, y = sv_framework_2_feature_2, color = color)
) +
geom_point(size = 1) +
xlab(paste(sv_framework_2_str, feature_1)) +
ylab(paste(sv_framework_2_str, feature_2)) +
scale_x_continuous(limits = c(-1500, 1000), breaks = c(-1000, 0, 1000)) +
scale_y_continuous(limits = c(-1000, 1000), breaks = c(-500, 0, 500)) +
scale_color_gradient(low = "blue", high = "red") +
theme_minimal() +
theme(
text = element_text(size = 12),
axis.text.x = element_text(size = 12),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank()
)
# Plot of the trend of the data
bike_plot_new <- ggplot(bike, aes(x = trend, y = cnt, color = get(color))) +
geom_point(size = 0.75) +
scale_color_gradient(low = "blue", high = "red") +
labs(color = color) +
xlab("Days since 1 January 2011") +
ylab("Number of bikes rented") +
theme_minimal() +
theme(legend.position = "right", legend.title = element_text(size = 10))
# Combine the plots
ggpubr::ggarrange(
bike_plot_new,
ggpubr::ggarrange(
scatterplot_topleft,
scatterplot_topright,
scatterplot_bottomleft,
scatterplot_bottomright,
legend = "none"
),
nrow = 2, heights = c(1, 2)
)
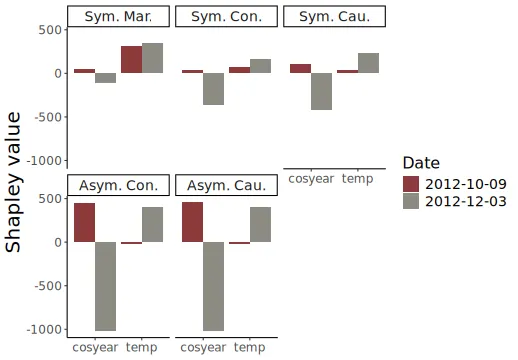
Investigating two similar days
We investigate the difference between symmetric/asymmetric conditional, symmetric/asymmetric causal, and marginal Shapley values for two days: October 10 and December 3, 2012. They have more or less the same temperature of 13 and 13.27 degrees Celsius, and predicted bike counts of 6117 and 6241, respectively. The figure below is an extension of Figure 4 in Heskes et al. (2020), as they only included asymmetric conditional, symmetric causal, and marginal Shapley values.
We plot the various Shapley values for the cosyear and
temp features below. We obtain the same results as Heskes et al. (2020) obtained, namely, that the
marginal Shapley value explanation framework provides similar
explanations for both days. I.e., it only considers the direct effect of
temp. The asymmetric conditional and causal Shapley values
are almost indistinguishable and put the most weight on the ‘root’ cause
cosyear. Heskes et al. (2020)
states that the symmetric causal Shapley values provide a sensible
balance between the two extremes and gives credit to both season and
temperature, but still different explanations for the two days.
However, as we also include symmetric conditional Shapley values, we see that they are extremely similar to symmetric causal Shapley values. I.e., the conditional Shapley value explanation framework also provides a sensible balance between marginal and asymmetric Shapley values. To summarize: as concluded by Heskes et al. (2020) in their Figure 4, the asymmetric conditional/causal Shapley values focus on the root cause, marginal Shapley values on the more direct effect, and symmetric conditional/causal Shapley values consider both for a more natural explanation.
# Features of interest
features <- c("cosyear", "temp")
# Get explicands with similar temperature: 2012-10-09 (October) and 2012-12-03 (December)
dates <- c("2012-10-09", "2012-12-03")
dates_idx <- sapply(dates, function(data) which(as.integer(row.names(x_explain)) == which(bike$dteday == data)))
# predict(model, x_explain)[dates_idx] + mean(y_train_nc) # predicted values for the two points
# List of the Shapley value explanations
explanations <- list(
"Sym. Mar." = explanation_sym_marg[["gaussian"]],
"Sym. Con." = explanation_sym_con[["gaussian"]],
"Sym. Cau." = explanation_sym_cau[["gaussian"]],
"Asym. Con." = explanation_asym_con[["gaussian"]],
"Asym. Cau." = explanation_asym_cau[["gaussian"]]
)
# Extract the relevant Shapley values
explanations_extracted <- data.table::rbindlist(lapply(seq_along(explanations), function(idx) {
explanations[[idx]]$shapley_values_est[
dates_idx, ..features
][, `:=`(Date = dates, type = names(explanations)[idx])]
}))
# Set type to be an ordered factor
explanations_extracted[, type := factor(type, levels = names(explanations), ordered = TRUE)]
# Convert from wide to long data table
dt_all <- data.table::melt(explanations_extracted,
id.vars = c("Date", "type"),
variable.name = "feature"
)
# Make the plot
ggplot(dt_all, aes(
x = feature, y = value, group = interaction(Date, feature),
fill = Date, label = round(value, 2)
)) +
geom_col(position = "dodge") +
theme_classic() +
ylab("Shapley value") +
facet_wrap(vars(type)) +
theme(axis.title.x = element_blank()) +
scale_fill_manual(values = c("indianred4", "ivory4")) +
theme(
legend.position.inside = c(0.75, 0.25), axis.title = element_text(size = 20),
legend.title = element_text(size = 16), legend.text = element_text(size = 14),
axis.text.x = element_text(size = 12), axis.text.y = element_text(size = 12),
strip.text.x = element_text(size = 14)
)
We can also make a similar plot using the
plot_SV_several_approaches function in shapr,
but then we get each explicand in a separate facet instead of a facet
for each framework.
# Here 2012-10-09 is the left facet and 2012-12-03 the right facet
plot_SV_several_approaches(explanations,
index_explicands = dates_idx,
only_these_features = features, # Can include more features.
facet_scales = "free_x",
horizontal_bars = FALSE,
axis_labels_n_dodge = 1,
print_ggplot = FALSE
) + theme(legend.position = "bottom")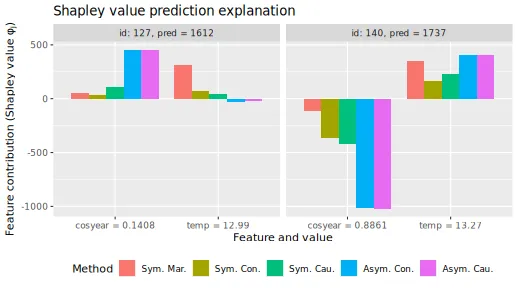
Furthermore, instead of doing as Heskes et al.
(2020) and only considering the features cosyear and
temp, we can plot all features, too, to get a more complete
overview.
# Here 2012-10-09 is the left facet and 2012-12-03 the right facet
plot_SV_several_approaches(explanations,
index_explicands = dates_idx,
facet_scales = "free_x",
horizontal_bars = FALSE,
axis_labels_rotate_angle = 45,
digits = 2,
print_ggplot = FALSE
) + theme(legend.position = "bottom")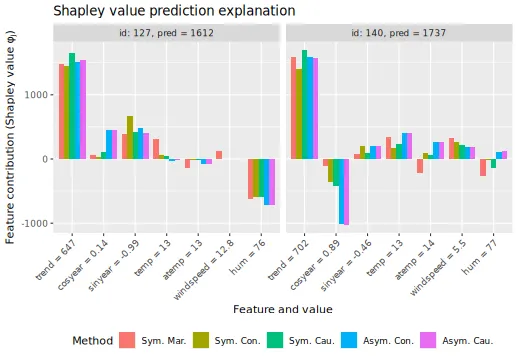
Sampling of coalitions
We can use max_n_coalitions to specify/reduce the number
of coalitions to use when computing the Shapley value explanation
framework. This applies to marginal, conditional, and causal Shapley
values, both the symmetric and asymmetric versions. However, recall that
the asymmetric versions already have fewer valid coalitions due to the
causal ordering.
In the example below, we demonstrate the sampling of coalitions for the asymmetric and symmetric causal Shapley value explanation frameworks. We half the number of coalitions for both versions and see that the elapsed times are approximately halved, too.
explanation_n_coal <- list()
explanation_n_coal[["sym_cau_gaussian_64"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
approach = "gaussian",
phi0 = phi0,
seed = 1,
asymmetric = FALSE,
causal_ordering = list(1, 2:3, 4:7),
confounding = c(FALSE, TRUE, FALSE),
max_n_coalitions = 64 # Instead of 128
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:07:01
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: gaussian
#>
#> • Procedure: Iterative
#>
#> • Number of Monte Carlo integration samples: 1000
#>
#> • Number of feature-wise Shapley values: 7
#>
#> • Number of observations to explain: 144
#>
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#>
#> • Components with confounding: {cosyear, sinyear}
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16487c063765.rds'
#>
#>
#>
#> ── Iterative computation started ──
#>
#>
#>
#> ── Iteration 1 ───────────────────────────────────────
#>
#> ℹ Using 14 of 128 coalitions, 14 new.
#>
#>
#>
#> ── Iteration 2 ───────────────────────────────────────
#>
#> ℹ Using 26 of 128 coalitions, 12 new.
#>
#>
#>
#> ── Iteration 3 ───────────────────────────────────────
#>
#> ℹ Using 46 of 128 coalitions, 20 new.
#>
#>
#>
#> ── Iteration 4 ───────────────────────────────────────
#>
#> ℹ Using 50 of 128 coalitions, 4 new.
#>
#>
#>
#> ── Iteration 5 ───────────────────────────────────────
#>
#> ℹ Using 52 of 128 coalitions, 2 new.
explanation_n_coal[["asym_cau_gaussian_10"]] <- explain(
model = model,
x_train = x_train,
x_explain = x_explain,
approach = "gaussian",
phi0 = phi0,
seed = 1,
asymmetric = TRUE,
causal_ordering = list(1, 2:3, 4:7),
confounding = c(FALSE, TRUE, FALSE),
verbose = c("basic", "convergence", "shapley"),
max_n_coalitions = 10, # Instead of 20
iterative = FALSE # Due to small number of coalitions
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:07:18
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 7
#> • Number of observations to explain: 144
#> • Number of asymmetric coalitions: 20
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp, atemp, windspeed, hum}
#> • Components with confounding: {cosyear, sinyear}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16481ee959ab.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 10 of 10 coalitions.
#>
#> ── Convergence info
#> ✔ Iterative Shapley value estimation stopped at 10 coalitions after 1 iterations, due to:
#> Maximum number of iterations (1) reached!
#> Maximum number of coalitions (10) reached!
#>
#> Final estimated Shapley values (sd)
#> explain_id none trend cosyear sinyear
#> <int> <char> <char> <char> <char>
#> 1: 1 0 (0) -1973.19 ( 298.81) -974.81 ( 310.79) -335.07 (250.87)
#> 2: 2 0 (0) -1975.59 ( 302.82) -967.43 ( 319.35) -290.53 (264.71)
#> 3: 3 0 (0) -1930.76 ( 296.16) -973.40 ( 322.03) -186.42 (261.74)
#> 4: 4 0 (0) -1932.90 ( 301.60) -958.90 ( 330.38) -149.19 (274.88)
#> 5: 5 0 (0) -1877.93 ( 305.06) -852.85 ( 314.05) -207.57 (239.95)
#> ---
#> 140: 140 0 (0) 995.51 ( 362.01) -479.45 ( 325.45) 187.74 (253.89)
#> 141: 141 0 (0) 1012.39 ( 381.91) -530.82 ( 333.78) 42.14 (135.80)
#> 142: 142 0 (0) 1012.41 ( 425.56) -702.60 ( 371.05) -97.43 (159.67)
#> 143: 143 0 (0) 1019.65 ( 731.08) -2022.93 ( 676.61) -304.02 (368.61)
#> 144: 144 0 (0) 1016.02 (1025.42) -3328.06 (1003.39) -268.07 (635.13)
#> temp atemp windspeed hum
#> <char> <char> <char> <char>
#> 1: -37.48 ( 65.72) -20.18 ( 55.05) 121.97 ( 73.83) 11.33 ( 94.52)
#> 2: -28.41 ( 60.78) -3.86 ( 29.06) 17.47 ( 46.76) 205.18 ( 94.06)
#> 3: -31.78 ( 69.57) -4.72 ( 32.51) 21.73 ( 28.34) -331.86 ( 72.67)
#> 4: 65.18 ( 43.48) 16.32 ( 28.82) -112.62 ( 30.50) -88.37 ( 56.64)
#> 5: 40.33 ( 48.58) 12.35 ( 22.21) 110.09 ( 29.35) -137.02 ( 54.02)
#> ---
#> 140: 61.56 (168.08) 98.78 (168.21) 594.81 (147.78) 533.16 (210.24)
#> 141: -21.32 ( 43.50) 26.16 ( 22.80) 176.34 ( 22.24) -190.41 ( 53.27)
#> 142: -26.81 ( 68.60) 87.47 ( 26.02) -32.60 ( 31.01) 279.85 ( 87.32)
#> 143: -16.21 ( 54.12) -18.45 ( 41.76) -72.97 ( 53.34) -65.21 ( 97.54)
#> 144: 14.35 (131.34) -28.08 ( 67.88) -145.59 ( 68.02) -528.08 (161.40)
# Look at the times
explanation_n_coal[["sym_cau_gaussian_all_128"]] <- explanation_sym_cau$gaussian
explanation_n_coal[["asym_cau_gaussian_all_20"]] <- explanation_asym_cau$gaussian
explanation_n_coal <- explanation_n_coal[c(1, 3, 2, 4)]
print_time(explanation_n_coal)
#> sym_cau_gaussian_64 sym_cau_gaussian_all_128 asym_cau_gaussian_10
#> [1,] 16.486 35.7 2.2132
#> asym_cau_gaussian_all_20
#> [1,] 5.4413We can then plot the beeswarm plots and the Shapley values for the six selected explicands. We see that there are only minuscule differences between the Shapley values we obtain when we use all the coalitions and those we obtain when we use half of the valid coalitions.
plot_beeswarms(explanation_n_coal, title = "Shapley values (gaussian) exact vs. approximation")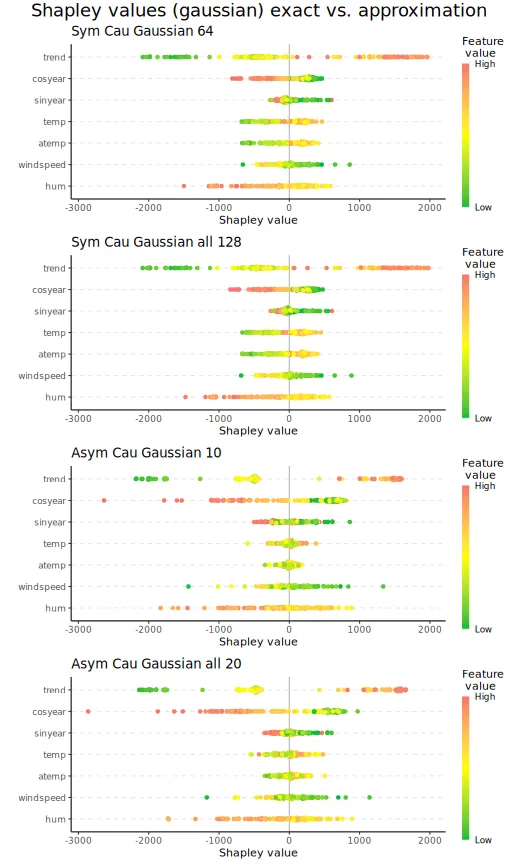
plot_SV_several_approaches(explanation_n_coal, index_x_explain, print_ggplot = FALSE) +
theme(legend.position = "bottom") +
guides(fill = guide_legend(nrow = 2))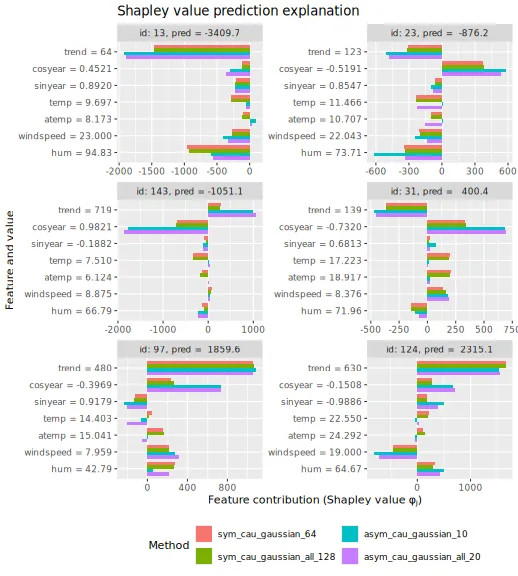
Groups of features
In this section, we demonstrate that we can compute marginal,
asymmetric conditional, and symmetric/asymmetric Shapley values for
groups of features, too. For group Shapley values, we need to specify
the causal ordering on the group level and the feature level. We
demonstrate with the gaussian approach, but other
approaches are applicable, too.
In the pairs plot above (and below), we see that it can be natural to
group the features temp and atemp due to their
(conceptual) similarity and high correlation.
GGally::ggpairs(x_train[, 4:5])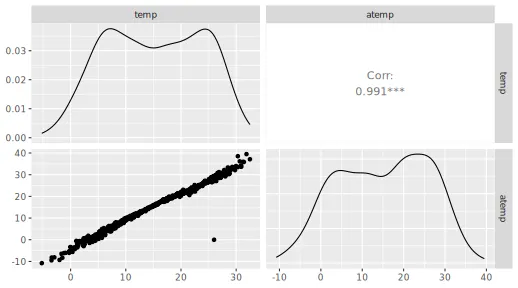
We set up the groups and update the causal ordering to be on the group level.
group_list <- list(
trend = "trend",
cosyear = "cosyear",
sinyear = "sinyear",
temp_group = c("temp", "atemp"),
windspeed = "windspeed",
hum = "hum"
)
causal_ordering_group <-
list("trend", c("cosyear", "sinyear"), c("temp_group", "windspeed", "hum"))
confounding <- c(FALSE, TRUE, FALSE)We can then compute the (group) Shapley values using the different Shapley value frameworks.
explanation_group_gaussian <- list()
explanation_group_gaussian[["symmetric_marginal"]] <-
explain(
model = model,
x_train = x_train,
x_explain = x_explain,
approach = "gaussian",
phi0 = phi0,
seed = 1,
asymmetric = FALSE,
causal_ordering = list(seq(length(group_list))), # or `NULL`
confounding = TRUE,
n_MC_samples = 1000,
group = group_list,
iterative = FALSE
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:07:23
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_groups = 64`, and is therefore set to
#> `2^n_groups = 64`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgboost>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: gaussian
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 1000
#>
#> • Number of group-wise Shapley values: 6
#>
#> • Feature groups: trend: {"trend"}; cosyear:
#> {"cosyear"}; sinyear: {"sinyear"}; temp_group:
#> {"temp", "atemp"}; windspeed: {"windspeed"}; hum:
#> {"hum"}
#>
#> • Number of observations to explain: 144
#>
#> • Causal ordering: {trend, cosyear, sinyear,
#> temp_group, windspeed, hum}
#>
#> • Components with confounding: {trend, cosyear,
#> sinyear, temp_group, windspeed, hum}
#>
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16489e235da.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 64 of 64 coalitions.
explanation_group_gaussian[["symmetric_conditional"]] <-
explain(
model = model,
x_train = x_train,
x_explain = x_explain,
approach = "gaussian",
phi0 = phi0,
seed = 1,
asymmetric = FALSE,
causal_ordering = list(seq(length(group_list))), # or `NULL`
confounding = NULL,
n_MC_samples = 1000,
group = group_list,
iterative = FALSE
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:07:31
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_groups = 64`, and is therefore set to
#> `2^n_groups = 64`.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of group-wise Shapley values: 6
#> • Feature groups: trend: {"trend"}; cosyear:
#> {"cosyear"}; sinyear: {"sinyear"}; temp_group:
#> {"temp", "atemp"}; windspeed: {"windspeed"}; hum:
#> {"hum"}
#> • Number of observations to explain: 144
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164846c998ae.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 64 of 64 coalitions.
explanation_group_gaussian[["asymmetric_conditional"]] <-
explain(
model = model,
x_train = x_train,
x_explain = x_explain,
approach = "gaussian",
phi0 = phi0,
seed = 1,
asymmetric = TRUE,
causal_ordering = causal_ordering_group,
confounding = NULL,
n_MC_samples = 1000,
group = group_list,
iterative = FALSE
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:07:37
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than the
#> number of coalitions respecting the causal ordering
#> (12), and is therefore set to 12.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of group-wise Shapley values: 6
#> • Feature groups: trend: {"trend"}; cosyear:
#> {"cosyear"}; sinyear: {"sinyear"}; temp_group:
#> {"temp", "atemp"}; windspeed: {"windspeed"}; hum:
#> {"hum"}
#> • Number of observations to explain: 144
#> • Number of asymmetric coalitions: 12
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp_group, windspeed, hum}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_16481145a886.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 12 of 12 coalitions.
explanation_group_gaussian[["symmetric_causal"]] <-
explain(
model = model,
x_train = x_train,
x_explain = x_explain,
approach = "gaussian",
phi0 = phi0,
seed = 1,
asymmetric = FALSE,
causal_ordering = causal_ordering_group,
confounding = confounding,
n_MC_samples = 1000,
group = group_list,
iterative = FALSE
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:07:38
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than
#> `2^n_groups = 64`, and is therefore set to
#> `2^n_groups = 64`.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of group-wise Shapley values: 6
#> • Feature groups: trend: {"trend"}; cosyear:
#> {"cosyear"}; sinyear: {"sinyear"}; temp_group:
#> {"temp", "atemp"}; windspeed: {"windspeed"}; hum:
#> {"hum"}
#> • Number of observations to explain: 144
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp_group, windspeed, hum}
#> • Components with confounding: {cosyear, sinyear}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_164879eeea7e.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 64 of 64 coalitions.
explanation_group_gaussian[["asymmetric_causal"]] <-
explain(
model = model,
x_train = x_train,
x_explain = x_explain,
approach = "gaussian",
phi0 = phi0,
seed = 1,
asymmetric = TRUE,
causal_ordering = causal_ordering_group,
confounding = confounding,
n_MC_samples = 1000,
group = group_list,
iterative = FALSE
)
#>
#> ── Starting `shapr::explain()` at 2026-01-16 13:07:57
#> ℹ Feature classes extracted from the model contain
#> `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than the
#> number of coalitions respecting the causal ordering
#> (12), and is therefore set to 12.
#> ── Explanation overview ──
#>
#> • Model class: <xgboost>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of group-wise Shapley values: 6
#> • Feature groups: trend: {"trend"}; cosyear:
#> {"cosyear"}; sinyear: {"sinyear"}; temp_group:
#> {"temp", "atemp"}; windspeed: {"windspeed"}; hum:
#> {"hum"}
#> • Number of observations to explain: 144
#> • Number of asymmetric coalitions: 12
#> • Causal ordering: {trend}, {cosyear, sinyear},
#> {temp_group, windspeed, hum}
#> • Components with confounding: {cosyear, sinyear}
#> • Computations (temporary) saved at:
#> '/tmp/Rtmpravdkd/shapr_obj_1648465b5494.rds'
#>
#> ── Main computation started ──
#>
#> ℹ Using 12 of 12 coalitions.
# Look at the elapsed times (symmetric takes the longest time)
print_time(explanation_group_gaussian)
#> symmetric_marginal symmetric_conditional asymmetric_conditional
#> [1,] 8.7174 5.2062 1.2685
#> symmetric_causal asymmetric_causal
#> [1,] 18.989 2.5814We can then make the beeswarm plots and Shapley value plots for the
six selected explicands. For the beeswarm plots, we set
include_group_feature_means = TRUE to make the plots. This
means that the plot function uses the mean of the temp and
atemp features as the feature value. This only makes sense
due to the high correlation between the two features.
The main difference between the feature-wise and group-wise Shapley
values is that we now see a much wider spread in the Shapley values for
temp_group than we did for temp and
atemp. For example, for the symmetric causal framework, we
saw above that the temp and atemp obtained
Shapley values between (around)
and
,
while the grouped version temp_group obtains Shapley values
between
and
.
plot_beeswarms(explanation_group_gaussian,
title = "Group Shapley values (gaussian)",
include_group_feature_means = TRUE
)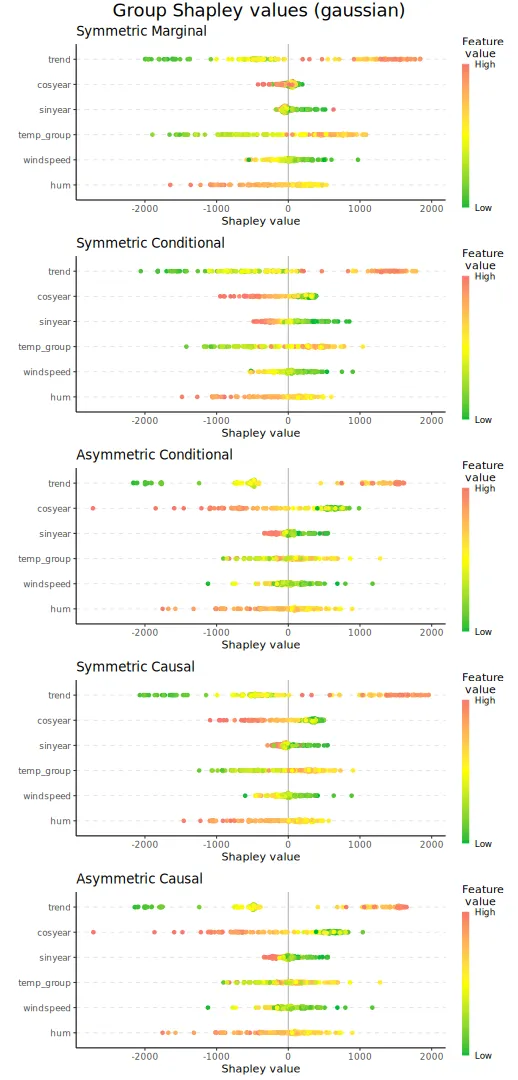
plot_SV_several_approaches(explanation_group_gaussian, index_x_explain, print_ggplot = FALSE) +
ggtitle("Shapley value prediction explanation (gaussian)") +
theme(legend.position = "bottom") + guides(fill = guide_legend(nrow = 2))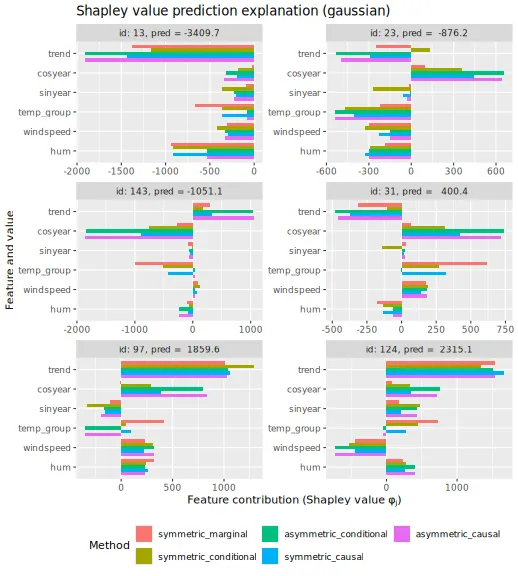
Implementation details
The shapr package is built to estimate conditional
Shapley values, thus, it parallelizes over the coalitions. This makes
perfect sense for said framework as each batch of coalitions is
independent of other batches, which means that it is easy to
parallelize. Furthermore, by using many batches, we drastically reduce
the memory usage as shapr does not need to store the Monte
Carlo samples for all coalitions.
This setup is not optimal for the causal Shapley value framework as the chains of sampling steps for two coalitions and can contain many of the same steps. Ideally, each unique sampling step should only be modeled once to save computation time, but some of the sampling steps will occur in many of the chains. Thus, we would then have to store the Monte Carlo samples for all coalitions where this sampling step is included, and we can therefore run into memory consumption problems. Thus, in the current implementation, we treat each coalition independently and remodel the needed sampling steps for each coalition.
Furthermore, in the conditional Shapley value framework, we have that
,
thus shapr will by default generate Monte Carlo samples for
all features not in
.
For the causal Shapley value framework, this is not the case, i.e.,
in general. To reuse the code, we generate Monte Carlo samples for all
features not in
,
but only keep the samples for the features in
.
To speed up shapr further, one could rewrite all the
approaches to support cases where
is not the complement of
.
In the code below, we see the unique coalitions/set of features to
condition on to generate the Monte Carlo samples for all coalitions and
the number of times that set of conditional features is needed in the
symmetric causal Shapley value framework for the setup above. We see
that most of the conditional distributions will now be remodeled eight
times. For the gaussian approach, which is very fast to
estimate the conditional distributions, this does not have a major
impact on the time. However, for, e.g., the ctree approach
which is much slower, this will take a significant amount of extra time.
The vaeac approach trains only on these relevant
coalitions.
S_causal_steps <- explanation_sym_cau$gaussian$internal$iter_list[[1]]$S_causal_steps
S_causal_unlist <- do.call(c, unlist(S_causal_steps, recursive = FALSE))
S_causal_steps_freq <- S_causal_unlist[grepl("\\.S(?!bar)", names(S_causal_unlist), perl = TRUE)]
S_causal_steps_freq <- S_causal_steps_freq[!sapply(S_causal_steps_freq, is.null)] # Remove NULLs
S_causal_steps_freq <- S_causal_steps_freq[sapply(S_causal_steps_freq, length) > 0] # Remove extra integer(0)
table(sapply(S_causal_steps_freq, paste0, collapse = ","))
#>
#> 1 1,2,3 1,2,3,4 1,2,3,4,5 1,2,3,4,5,6 1,2,3,4,5,7
#> 95 7 8 8 8 8
#> 1,2,3,4,6 1,2,3,4,6,7 1,2,3,4,7 1,2,3,5 1,2,3,5,6 1,2,3,5,6,7
#> 8 8 8 8 8 8
#> 1,2,3,5,7 1,2,3,6 1,2,3,6,7 1,2,3,7
#> 8 8 8 8The independence, empirical,
ctree, and categorical approaches produce
weighted Monte Carlo samples. That means that they do not necessarily
generate n_MC_samples. To ensure n_MC_samples,
we sample n_MC_samples samples using weighted sampling with
replacement where the weights are the weights returned by the
approaches.
The marginal Shapley value explanation framework can be extended to
support modeling the marginal distributions using the
copula and vaeac approaches as both of these
methods support unconditional sampling.