Explain the Output of Machine Learning Models with Dependence-Aware (Conditional/Observational) Shapley Values
Source:R/explain.R
explain.RdCompute dependence-aware Shapley values for observations in x_explain from the specified
model using the method specified in approach to estimate the conditional expectation.
See Aas et al. (2021)
for a thorough introduction to dependence-aware prediction explanation with Shapley values.
For an overview of the methodology and capabilities of the package, see the software paper
Jullum et al. (2025), or the pkgdown site at
norskregnesentral.github.io/shapr/.
Usage
explain(
model,
x_explain,
x_train,
approach,
phi0,
iterative = NULL,
max_n_coalitions = NULL,
group = NULL,
n_MC_samples = 1000,
seed = NULL,
verbose = "basic",
predict_model = NULL,
get_model_specs = NULL,
prev_shapr_object = NULL,
asymmetric = FALSE,
causal_ordering = NULL,
confounding = NULL,
extra_computation_args = list(),
iterative_args = list(),
output_args = list(),
...
)Arguments
- model
Model object. The model whose predictions you want to explain. Run
get_supported_models()for a table of which modelsexplainsupports natively. Unsupported models can still be explained by passingpredict_modeland (optionally)get_model_specs, see details for more information.- x_explain
Matrix or data.frame/data.table. Features for which predictions should be explained.
- x_train
Matrix or data.frame/data.table. Data used to estimate the (conditional) feature distributions needed to properly estimate the conditional expectations in the Shapley formula.
- approach
Character vector of length
1or one less than the number of features. All elements should either be"gaussian","copula","empirical","ctree","vaeac","categorical","timeseries","independence","regression_separate", or"regression_surrogate". The two regression approaches cannot be combined with any other approach. See details for more information.- phi0
Numeric. The prediction value for unseen data, i.e., an estimate of the expected prediction without conditioning on any features. Typically set this equal to the mean of the response in the training data, but alternatives such as the mean of the training predictions are also reasonable.
- iterative
Logical or NULL. If
NULL(default), set toTRUEif there are more than 5 features/groups, andFALSEotherwise. IfTRUE, Shapley values are estimated iteratively for faster, sufficiently accurate results. First an initial number of coalitions is sampled, then bootstrapping estimates the variance of the Shapley values. A convergence criterion determines if the variances are sufficiently small. If not, additional samples are added. The process repeats until the variances are below the threshold. Specifics for the iterative process and convergence criterion are set viaiterative_args.- max_n_coalitions
Integer. Upper limit on the number of unique feature/group coalitions to use in the iterative procedure (if
iterative = TRUE). Ifiterative = FALSE, it represents the number of feature/group coalitions to use directly. The quantity refers to the number of unique feature coalitions ifgroup = NULL, and group coalitions ifgroup != NULL.max_n_coalitions = NULLcorresponds to2^n_features.- group
List. If
NULL, regular feature-wise Shapley values are computed. If provided, group-wise Shapley values are computed.groupthen has length equal to the number of groups. Each list element contains the character vectors with the features included in the corresponding group. See Jullum et al. (2021) for more information on group-wise Shapley values.- n_MC_samples
Positive integer. For most approaches, it indicates the maximum number of samples to use in the Monte Carlo integration of every conditional expectation. For
approach="ctree",n_MC_samplescorresponds to the number of samples from the leaf node (see an exception related to thectree.sampleargument insetup_approach.ctree()). Forapproach="empirical",n_MC_samplesis the \(K\) parameter in equations (14-15) of Aas et al. (2021), i.e. the maximum number of observations (with largest weights) that is used, see also theempirical.etaargumentsetup_approach.empirical().- seed
Positive integer. Specifies the seed before any code involving randomness is run. If
NULL(default), no seed is set in the calling environment.- verbose
String vector or NULL. Controls verbosity (printout detail level) via one or more of
"basic","progress","convergence","shapley"and"vS_details"."basic"(default) displays basic information about the computation and messages about parameters/checks."progress"displays where in the calculation process the function currently is."convergence"displays how close the Shapley value estimates are to convergence (only wheniterative = TRUE)."shapley"displays intermediate Shapley value estimates and standard deviations (only wheniterative = TRUE), and the final estimates."vS_details"displays information about the v(S) estimates, most relevant forapproach %in% c("regression_separate", "regression_surrogate", "vaeac").NULLmeans no printout. Any combination can be used, e.g.,verbose = c("basic", "vS_details").- predict_model
Function. Prediction function to use when
modelis not natively supported. (Runget_supported_models()for a list of natively supported models.) The function must have two arguments,modelandnewdata, which specify the model and a data.frame/data.table to compute predictions for, respectively. The function must give the prediction as a numeric vector.NULL(the default) uses functions specified internally. Can also be used to override the default function for natively supported model classes.- get_model_specs
Function. An optional function for checking model/data consistency when
modelis not natively supported. (Runget_supported_models()for a list of natively supported models.) The function takesmodelas an argument and provides a list with 3 elements:- labels
Character vector with the names of each feature.
- classes
Character vector with the class of each feature.
- factor_levels
Character vector with the levels for any categorical features.
If
NULL(the default), internal functions are used for natively supported model classes, and checking is disabled for unsupported model classes. Can also be used to override the default function for natively supported model classes.- prev_shapr_object
shaprobject or string. If an object of classshapris provided, or a string with a path to where intermediate results are stored, then the function will use the previous object to continue the computation. This is useful if the computation is interrupted or you want higher accuracy than already obtained, and therefore want to continue the iterative estimation. See the general usage vignette for examples.- asymmetric
Logical. Not applicable for (regular) non-causal explanations. If
FALSE(default),explaincomputes regular symmetric Shapley values. IfTRUE,explaincomputes asymmetric Shapley values based on the (partial) causal ordering given bycausal_ordering. That is,explainonly uses feature coalitions that respect the causal ordering. IfasymmetricisTRUEandconfoundingisNULL(default),explaincomputes asymmetric conditional Shapley values as specified in Frye et al. (2020). Ifconfoundingis provided, i.e., notNULL, thenexplaincomputes asymmetric causal Shapley values as specified in Heskes et al. (2020).- causal_ordering
List. Not applicable for (regular) non-causal or asymmetric explanations.
causal_orderingis an unnamed list of vectors specifying the components of the partial causal ordering that the coalitions must respect. Each vector represents a component and contains one or more features/groups identified by their names (strings) or indices (integers). Ifcausal_orderingisNULL(default), no causal ordering is assumed and all possible coalitions are allowed. No causal ordering is equivalent to a causal ordering with a single component that includes all features (list(1:n_features)) or groups (list(1:n_groups)) for feature-wise and group-wise Shapley values, respectively. For feature-wise Shapley values andcausal_ordering = list(c(1, 2), c(3, 4)), the interpretation is that features 1 and 2 are the ancestors of features 3 and 4, while features 3 and 4 are on the same level. Note: All features/groups must be included incausal_orderingwithout duplicates.- confounding
Logical vector. Not applicable for (regular) non-causal or asymmetric explanations.
confoundingis a logical vector specifying whether confounding is assumed for each component in thecausal_ordering. IfNULL(default), no assumption about the confounding structure is made andexplaincomputes asymmetric/symmetric conditional Shapley values, depending onasymmetric. Ifconfoundingis a single logical (FALSEorTRUE), the assumption is set globally for all components in the causal ordering. Otherwise,confoundingmust have the same length ascausal_ordering, indicating the confounding assumption for each component. Whenconfoundingis specified,explaincomputes asymmetric/symmetric causal Shapley values, depending onasymmetric. Theapproachcannot beregression_separateorregression_surrogate, as the regression-based approaches are not applicable to the causal Shapley methodology.- extra_computation_args
Named list. Specifies extra arguments related to the computation of the Shapley values. See the help file of
get_extra_comp_args_default()for description of the arguments and their default values.- iterative_args
Named list. Specifies the arguments for the iterative procedure. See the help file of
get_iterative_args_default()for description of the arguments and their default values.- output_args
Named list. Specifies certain arguments related to the output of the function. See the help file of
get_output_args_default()for description of the arguments and their default values.- ...
Arguments passed on to
setup_approach.categorical,setup_approach.copula,setup_approach.ctree,setup_approach.empirical,setup_approach.gaussian,setup_approach.independence,setup_approach.regression_separate,setup_approach.regression_surrogate,setup_approach.timeseries,setup_approach.vaeaccategorical.joint_prob_dtData.table. (Optional) Containing the joint probability distribution for each combination of feature values.
NULLmeans it is estimated from thex_trainandx_explain.categorical.epsilonNumeric value. (Optional) If
categorical.joint_prob_dtis not supplied, probabilities/frequencies are estimated usingx_train. If certain observations occur inx_explainand NOT inx_train, then epsilon is used as the proportion of times that these observations occur in the training data. In theory, this proportion should be zero, but this causes an error later in the Shapley computation.internalList. Not used directly, but passed through from
explain().ctree.mincriterionNumeric scalar or vector. Either a scalar or vector of length equal to the number of features in the model. The value is equal to 1 - \(\alpha\) where \(\alpha\) is the nominal level of the conditional independence tests. If it is a vector, this indicates which value to use when conditioning on various numbers of features. The default value is 0.95.
ctree.minsplitNumeric scalar. Determines the minimum value that the sum of the left and right daughter nodes must reach for a split. The default value is 20.
ctree.minbucketNumeric scalar. Determines the minimum sum of weights in a terminal node required for a split. The default value is 7.
ctree.sampleBoolean. If
TRUE(default), then the method always samplesn_MC_samplesobservations from the leaf nodes (with replacement). IfFALSEand the number of observations in the leaf node is less thann_MC_samples, the method will take all observations in the leaf. IfFALSEand the number of observations in the leaf node is more thann_MC_samples, the method will samplen_MC_samplesobservations (with replacement). This means that there will always be sampling in the leaf unlesssample = FALSEand the number of obs in the node is less thann_MC_samples.empirical.typeCharacter. Must be one of
"fixed_sigma"(default),"AICc_each_k","AICc_full"or"independence". Note:"empirical.type = independence"is deprecated; useapproach = "independence"instead."fixed_sigma"uses a fixed bandwidth (set throughempirical.fixed_sigma) in the kernel density estimation."AICc_each_k"and"AICc_full"optimize the bandwidth using the AICc criterion, with respectively one bandwidth per coalition size and one bandwidth for all coalition sizes.empirical.etaNumeric scalar. Needs to be
0 < empirical.eta <= 1. The default value is 0.95. Represents the minimum proportion of the total empirical weight that data samples should use. For example, ifempirical.eta = .8, we choose theKsamples with the largest weights so that the sum of the weights accounts for 80% of the total weight.empirical.etais the \(\eta\) parameter in equation (15) of Aas et al. (2021).empirical.fixed_sigmaPositive numeric scalar. The default value is 0.1. Represents the kernel bandwidth in the distance computation used when conditioning on all different coalitions. Only used when
empirical.type = "fixed_sigma"empirical.n_samples_aiccPositive integer. Number of samples to consider in AICc optimization. The default value is 1000. Only used when
empirical.typeis either"AICc_each_k"or"AICc_full".empirical.eval_max_aiccPositive integer. Maximum number of iterations when optimizing the AICc. The default value is 20. Only used when
empirical.typeis either"AICc_each_k"or"AICc_full".empirical.start_aiccNumeric. Start value of the
sigmaparameter when optimizing the AICc. The default value is 0.1. Only used whenempirical.typeis either"AICc_each_k"or"AICc_full".empirical.cov_matNumeric matrix. The covariance matrix of the data generating distribution used to define the Mahalanobis distance.
NULLmeans it is estimated fromx_train.gaussian.muNumeric vector. Containing the mean of the data generating distribution.
NULLmeans it is estimated from thex_train.gaussian.cov_matNumeric matrix. Containing the covariance matrix of the data generating distribution.
NULLmeans it is estimated from thex_train.regression.modelA
tidymodelsobject of classmodel_specs. Default is a linear regression model, i.e.,parsnip::linear_reg(). See tidymodels for all possible models, and see the vignette for how to add new/own models. Note, to make it easier to callexplain()from Python, theregression.modelparameter can also be a string specifying the model which will be parsed and evaluated. For example,"parsnip::rand_forest(mtry = hardhat::tune(), trees = 100, engine = "ranger", mode = "regression")"is also a valid input. It is essential to include the package prefix if the package is not loaded.regression.tune_valuesEither
NULL(default), a data.frame/data.table/tibble, or a function. The data.frame must contain the possible hyperparameter value combinations to try. The column names must match the names of the tunable parameters specified inregression.model. Ifregression.tune_valuesis a function, then it should take one argumentxwhich is the training data for the current coalition and returns a data.frame/data.table/tibble with the properties described above. Using a function allows the hyperparameter values to change based on the size of the coalition See the regression vignette for several examples. Note, to make it easier to callexplain()from Python, theregression.tune_valuescan also be a string containing an R function. For example,"function(x) return(dials::grid_regular(dials::mtry(c(1, ncol(x)))), levels = 3))"is also a valid input. It is essential to include the package prefix if the package is not loaded.regression.vfold_cv_paraEither
NULL(default) or a named list containing the parameters to be sent torsample::vfold_cv(). See the regression vignette for several examples.regression.recipe_funcEither
NULL(default) or a function that that takes in arecipes::recipe()object and returns a modifiedrecipes::recipe()with potentially additional recipe steps. See the regression vignette for several examples. Note, to make it easier to callexplain()from Python, theregression.recipe_funccan also be a string containing an R function. For example,"function(recipe) return(recipes::step_ns(recipe, recipes::all_numeric_predictors(), deg_free = 2))"is also a valid input. It is essential to include the package prefix if the package is not loaded.regression.surrogate_n_combPositive integer. Specifies the number of unique coalitions to apply to each training observation. The default is the number of sampled coalitions in the present iteration. Any integer between 1 and the default is allowed. Larger values requires more memory, but may improve the surrogate model. If the user sets a value lower than the maximum, we sample this amount of unique coalitions separately for each training observations. That is, on average, all coalitions should be equally trained.
timeseries.fixed_sigmaPositive numeric scalar. Represents the kernel bandwidth in the distance computation. The default value is 2.
timeseries.boundsNumeric vector of length two. Specifies the lower and upper bounds of the timeseries. The default is
c(NULL, NULL), i.e. no bounds. If one or both of these bounds are notNULL, we restrict the sampled time series to be between these bounds. This is useful if the underlying time series are scaled between 0 and 1, for example.vaeac.depthPositive integer (default is
3). The number of hidden layers in the neural networks of the masked encoder, full encoder, and decoder.vaeac.widthPositive integer (default is
32). The number of neurons in each hidden layer in the neural networks of the masked encoder, full encoder, and decoder.vaeac.latent_dimPositive integer (default is
8). The number of dimensions in the latent space.vaeac.lrPositive numeric (default is
0.001). The learning rate used in thetorch::optim_adam()optimizer.vaeac.activation_functionAn
torch::nn_module()representing an activation function such as, e.g.,torch::nn_relu()(default),torch::nn_leaky_relu(),torch::nn_selu(), ortorch::nn_sigmoid().vaeac.n_vaeacs_initializePositive integer (default is
4). The number of different vaeac models to initiate in the start. Pick the best performing one aftervaeac.extra_parameters$epochs_initiation_phaseepochs (default is2) and continue training that one.vaeac.epochsPositive integer (default is
100). The number of epochs to train the final vaeac model. This includesvaeac.extra_parameters$epochs_initiation_phase, where the default is2.vaeac.extra_parametersNamed list with extra parameters to the
vaeacapproach. Seevaeac_get_extra_para_default()for description of possible additional parameters and their default values.
Value
Object of class c("shapr", "list"). Contains the following items:
shapley_values_estdata.table with the estimated Shapley values with explained observation in the rows and features along the columns. The column
noneis the prediction not devoted to any of the features (given by the argumentphi0)shapley_values_sddata.table with the standard deviation of the Shapley values reflecting the uncertainty in the coalition sampling part of the kernelSHAP procedure. These are, by definition, 0 when all coalitions are used. Only present when
extra_computation_args$compute_sd=TRUE, which is the default wheniterative = TRUE.internalList with the different parameters, data, functions and other output used internally.
pred_explainNumeric vector with the predictions for the explained observations.
MSEvList with the values of the MSEv evaluation criterion for the approach. See the MSEv evaluation section in the general usage vignette for details.
timingList containing timing information for the different parts of the computation.
summarycontains the time stamps for the start and end time in addition to the total execution time.overall_timing_secsgives the time spent on different parts of the explanation computation.main_computation_timing_secsfurther decomposes the main computation time into different parts of the computation for each iteration of the iterative estimation routine, if used.
Details
The shapr package implements kernelSHAP estimation of dependence-aware Shapley values with
eight different Monte Carlo-based approaches for estimating the conditional distributions of the data.
These are all introduced in the
general usage vignette.
(From R: vignette("general_usage", package = "shapr")).
For an overview of the methodology and capabilities of the package, please also see the software paper
Jullum et al. (2025).
Moreover,
Aas et al. (2021)
gives a general introduction to dependence-aware Shapley values and the approaches "empirical",
"gaussian", "copula", and also discusses "independence".
Redelmeier et al. (2020) introduces the approach "ctree".
Olsen et al. (2022) introduces the "vaeac"
approach.
Approach "timeseries" is discussed in
Jullum et al. (2021).
shapr has also implemented two regression-based approaches "regression_separate" and "regression_surrogate",
as described in Olsen et al. (2024).
It is also possible to combine the different approaches, see the
general usage vignette for more information.
The package also supports the computation of causal and asymmetric Shapley values as introduced by
Heskes et al. (2020) and
Frye et al. (2020).
Asymmetric Shapley values were proposed by
Frye et al. (2020) as a way to incorporate causal knowledge in
the real world by restricting the possible feature combinations/coalitions when computing the Shapley values to
those consistent with a (partial) causal ordering.
Causal Shapley values were proposed by
Heskes et al. (2020) as a way to explain the total effect of features
on the prediction, taking into account their causal relationships, by adapting the sampling procedure in shapr.
The package allows parallelized computation with progress updates through the tightly connected
future::future and progressr::progressr packages.
See the examples below.
For iterative estimation (iterative=TRUE), intermediate results may be printed to the console
(according to the verbose argument).
Moreover, the intermediate results are written to disk.
This combined batch computation of the v(S) values enables fast and accurate estimation of the Shapley values
in a memory-friendly manner.
Examples
# \donttest{
# Load example data
data("airquality")
airquality <- airquality[complete.cases(airquality), ]
x_var <- c("Solar.R", "Wind", "Temp", "Month")
y_var <- "Ozone"
# Split data into test and training data
data_train <- head(airquality, -3)
data_explain <- tail(airquality, 3)
x_train <- data_train[, x_var]
x_explain <- data_explain[, x_var]
# Fit a linear model
lm_formula <- as.formula(paste0(y_var, " ~ ", paste0(x_var, collapse = " + ")))
model <- lm(lm_formula, data = data_train)
# Explain predictions
p <- mean(data_train[, y_var])
# (Optionally) enable parallelization via the future package
if (requireNamespace("future", quietly = TRUE)) {
future::plan("multisession", workers = 2)
}
# (Optionally) enable progress updates within every iteration via the progressr package
if (requireNamespace("progressr", quietly = TRUE)) {
progressr::handlers(global = TRUE)
}
#> Error in globalCallingHandlers(condition = global_progression_handler): should not be called with handlers on the stack
# Empirical approach
explain1 <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "empirical",
phi0 = p,
n_MC_samples = 1e2
)
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:04 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_features = 16`, and is
#> therefore set to `2^n_features = 16`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: empirical
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 100
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9ed101e36.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
# Gaussian approach
explain2 <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "gaussian",
phi0 = p,
n_MC_samples = 1e2
)
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:08 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_features = 16`, and is
#> therefore set to `2^n_features = 16`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 100
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e4465fc98.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
# Gaussian copula approach
explain3 <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "copula",
phi0 = p,
n_MC_samples = 1e2
)
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:09 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_features = 16`, and is
#> therefore set to `2^n_features = 16`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: copula
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 100
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e50b0bd8d.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
if (requireNamespace("party", quietly = TRUE)) {
# ctree approach
explain4 <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "ctree",
phi0 = p,
n_MC_samples = 1e2
)
}
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:11 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_features = 16`, and is
#> therefore set to `2^n_features = 16`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: ctree
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 100
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e1a5dcc31.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
# Combined approach
approach <- c("gaussian", "gaussian", "empirical")
explain5 <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = approach,
phi0 = p,
n_MC_samples = 1e2
)
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:14 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_features = 16`, and is
#> therefore set to `2^n_features = 16`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian, gaussian, and empirical
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 100
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e7929f6a.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
## Printing
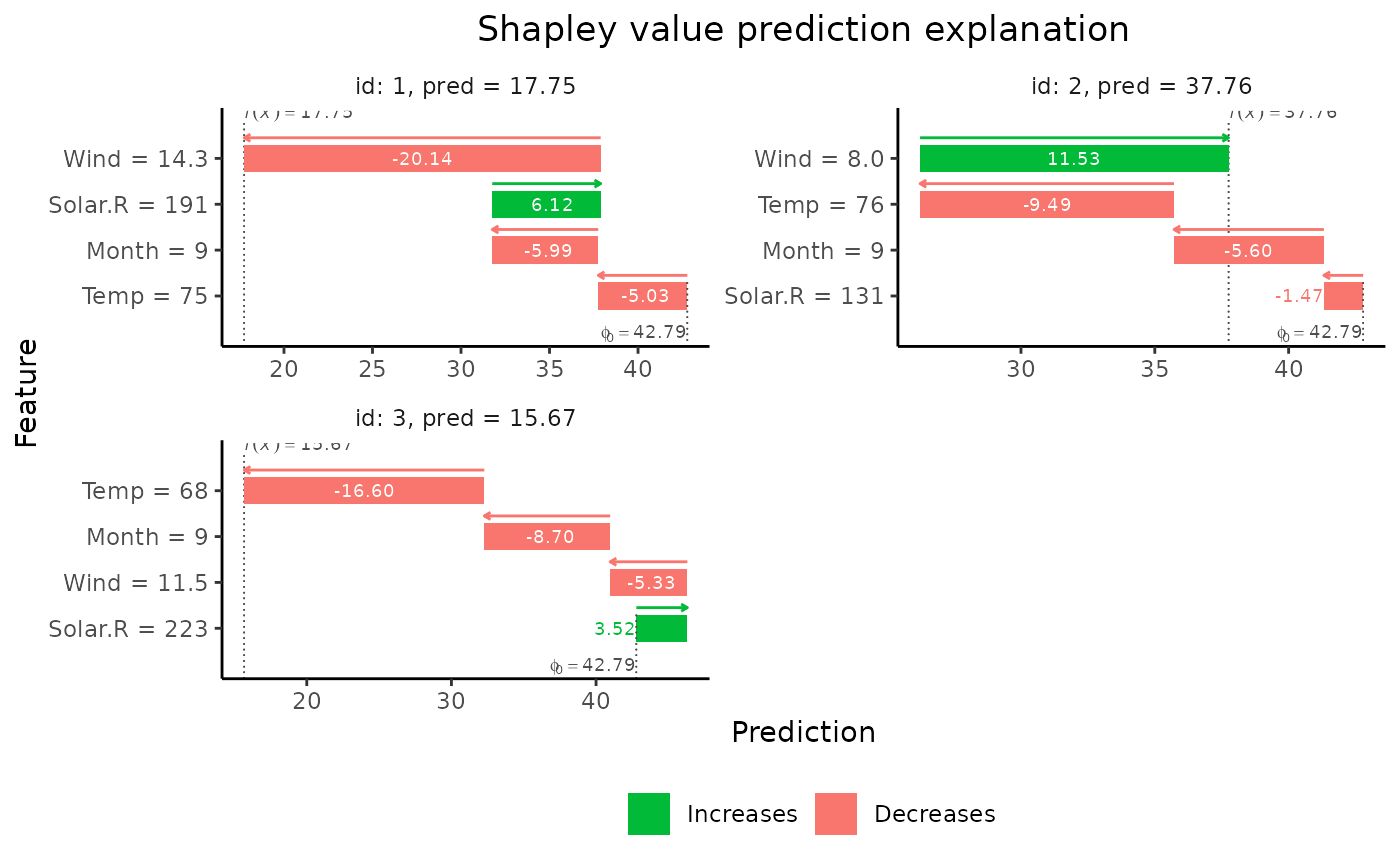
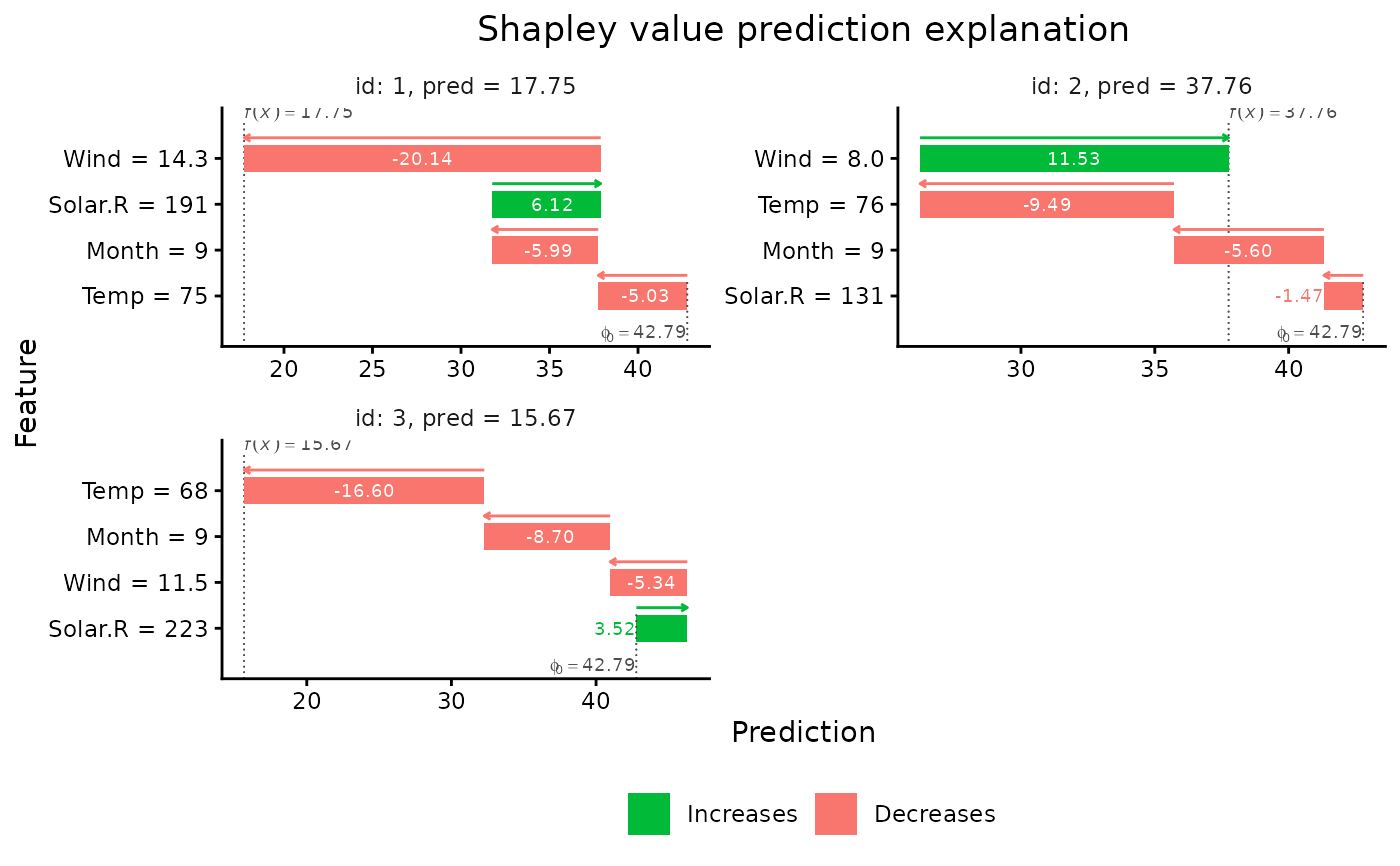
print(explain1) # The Shapley values
#> explain_id none Solar.R Wind Temp Month
#> <int> <num> <num> <num> <num> <num>
#> 1: 1 42.8 6.12 -20.14 -5.03 -5.99
#> 2: 2 42.8 -1.47 11.53 -9.49 -5.60
#> 3: 3 42.8 3.52 -5.34 -16.60 -8.70
print(explain1) # The Shapley values
#> explain_id none Solar.R Wind Temp Month
#> <int> <num> <num> <num> <num> <num>
#> 1: 1 42.8 6.12 -20.14 -5.03 -5.99
#> 2: 2 42.8 -1.47 11.53 -9.49 -5.60
#> 3: 3 42.8 3.52 -5.34 -16.60 -8.70
# The MSEv criterion (+sd). Smaller values indicate a better approach.
print(explain1, what = "MSEv")
#> MSEv MSEv_sd
#> <num> <num>
#> 1: 233 81.6
print(explain2, what = "MSEv")
#> MSEv MSEv_sd
#> <num> <num>
#> 1: 255 93.8
print(explain3, what = "MSEv")
#> MSEv MSEv_sd
#> <num> <num>
#> 1: 239 81.5
## Summary
summary1 <- summary(explain1)
summary1 # Provides a nicely formatted summary of the explanation
#>
#> ── Summary of Shapley value explanation ────────────────────────────────────────
#> • Computed with `shapr::explain()` in 3.3 seconds, started 2026-02-05 16:52:04
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: empirical
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 100
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Number of coalitions used: 16 (of total 16)
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9ed101e36.rds
#>
#> ── Estimated Shapley values
#> explain_id none Solar.R Wind Temp Month
#> <int> <char> <char> <char> <char> <char>
#> 1: 1 42.79 6.12 -20.14 -5.03 -5.99
#> 2: 2 42.79 -1.47 11.53 -9.49 -5.60
#> 3: 3 42.79 3.52 -5.34 -16.60 -8.70
#>
#> ── Estimated MSEv
#> Estimated MSE of v(S) = 233 (with sd = 82)
# Various additional info stored in the summary object
# Examples
summary1$shapley_est # A data.table with the Shapley values
#> explain_id none Solar.R Wind Temp Month
#> <int> <num> <num> <num> <num> <num>
#> 1: 1 42.78704 6.124296 -20.137653 -5.033967 -5.987303
#> 2: 2 42.78704 -1.470838 11.525868 -9.487924 -5.597657
#> 3: 3 42.78704 3.524599 -5.335059 -16.599988 -8.703929
summary1$timing$total_time_secs # Total computation time in seconds
#> NULL
summary1$parameters$n_MC_samples # Number of Monte Carlo samples used for the numerical integration
#> [1] 100
summary1$parameters$empirical.type # Type of empirical approach used
#> [1] "fixed_sigma"
# Plot the results
if (requireNamespace("ggplot2", quietly = TRUE)) {
plot(explain1)
plot(explain1, plot_type = "waterfall")
}
 # Group-wise explanations
group_list <- list(A = c("Temp", "Month"), B = c("Wind", "Solar.R"))
explain_groups <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
group = group_list,
approach = "empirical",
phi0 = p,
n_MC_samples = 1e2
)
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:18 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_groups = 4`, and is
#> therefore set to `2^n_groups = 4`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: empirical
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 100
#> • Number of group-wise Shapley values: 2
#> • Feature groups: A: {"Temp", "Month"}; B: {"Wind", "Solar.R"}
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e2f5c752a.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 4 of 4 coalitions.
print(explain_groups)
#> explain_id none A B
#> <int> <num> <num> <num>
#> 1: 1 42.8 -11.6 -13.40
#> 2: 2 42.8 -10.4 5.34
#> 3: 3 42.8 -25.8 -1.32
# Separate and surrogate regression approaches with linear regression models.
req_pkgs <- c("parsnip", "recipes", "workflows", "rsample", "tune", "yardstick")
if (requireNamespace(req_pkgs, quietly = TRUE)) {
explain_separate_lm <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
phi0 = p,
approach = "regression_separate",
regression.model = parsnip::linear_reg()
)
explain_surrogate_lm <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
phi0 = p,
approach = "regression_surrogate",
regression.model = parsnip::linear_reg()
)
}
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:21 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_features = 16`, and is
#> therefore set to `2^n_features = 16`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Regression
#> • Approach: regression_separate
#> • Procedure: Non-iterative
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e78b543ad.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:24 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_features = 16`, and is
#> therefore set to `2^n_features = 16`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Regression
#> • Approach: regression_surrogate
#> • Procedure: Non-iterative
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e156e6cc.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
# Iterative estimation
# For illustration only. By default not used for such small dimensions as here.
# Restricting the initial and maximum number of coalitions as well.
explain_iterative <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "gaussian",
phi0 = p,
iterative = TRUE,
iterative_args = list(initial_n_coalitions = 8),
max_n_coalitions = 12
)
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:26 ──────────────────────────
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian
#> • Procedure: Iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e146cfadd.rds
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ─────────────────────────────────────────────────────────────────
#> ℹ Using 8 of 16 coalitions, 8 new.
#>
#> ── Iteration 2 ─────────────────────────────────────────────────────────────────
#> ℹ Using 10 of 16 coalitions, 2 new.
#>
#> ── Iteration 3 ─────────────────────────────────────────────────────────────────
#> ℹ Using 12 of 16 coalitions, 2 new.
# When not using all coalitions, we can also get the SD of the Shapley values,
# reflecting uncertainty in the coalition sampling part of the procedure.
print(explain_iterative, what = "shapley_sd")
#> explain_id none Solar.R Wind Temp Month
#> <int> <num> <num> <num> <num> <num>
#> 1: 1 0 0.381 1.63 1.89 0.771
#> 2: 2 0 0.428 2.69 2.64 0.907
#> 3: 3 0 0.423 3.09 3.01 0.978
## Summary
# For iterative estimation, convergence info is also provided
summary_iterative <- summary(explain_iterative)
# }
# Group-wise explanations
group_list <- list(A = c("Temp", "Month"), B = c("Wind", "Solar.R"))
explain_groups <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
group = group_list,
approach = "empirical",
phi0 = p,
n_MC_samples = 1e2
)
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:18 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_groups = 4`, and is
#> therefore set to `2^n_groups = 4`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: empirical
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 100
#> • Number of group-wise Shapley values: 2
#> • Feature groups: A: {"Temp", "Month"}; B: {"Wind", "Solar.R"}
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e2f5c752a.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 4 of 4 coalitions.
print(explain_groups)
#> explain_id none A B
#> <int> <num> <num> <num>
#> 1: 1 42.8 -11.6 -13.40
#> 2: 2 42.8 -10.4 5.34
#> 3: 3 42.8 -25.8 -1.32
# Separate and surrogate regression approaches with linear regression models.
req_pkgs <- c("parsnip", "recipes", "workflows", "rsample", "tune", "yardstick")
if (requireNamespace(req_pkgs, quietly = TRUE)) {
explain_separate_lm <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
phi0 = p,
approach = "regression_separate",
regression.model = parsnip::linear_reg()
)
explain_surrogate_lm <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
phi0 = p,
approach = "regression_surrogate",
regression.model = parsnip::linear_reg()
)
}
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:21 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_features = 16`, and is
#> therefore set to `2^n_features = 16`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Regression
#> • Approach: regression_separate
#> • Procedure: Non-iterative
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e78b543ad.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:24 ──────────────────────────
#> ℹ `max_n_coalitions` is `NULL` or larger than `2^n_features = 16`, and is
#> therefore set to `2^n_features = 16`.
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Regression
#> • Approach: regression_surrogate
#> • Procedure: Non-iterative
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e156e6cc.rds
#>
#> ── Main computation started ──
#>
#> ℹ Using 16 of 16 coalitions.
# Iterative estimation
# For illustration only. By default not used for such small dimensions as here.
# Restricting the initial and maximum number of coalitions as well.
explain_iterative <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "gaussian",
phi0 = p,
iterative = TRUE,
iterative_args = list(initial_n_coalitions = 8),
max_n_coalitions = 12
)
#>
#> ── Starting `shapr::explain()` at 2026-02-05 16:52:26 ──────────────────────────
#>
#> ── Explanation overview ──
#>
#> • Model class: <lm>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: gaussian
#> • Procedure: Iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 3
#> • Computations (temporary) saved at: /tmp/RtmpWGjJWj/shapr_obj_1b9e146cfadd.rds
#>
#> ── Iterative computation started ──
#>
#> ── Iteration 1 ─────────────────────────────────────────────────────────────────
#> ℹ Using 8 of 16 coalitions, 8 new.
#>
#> ── Iteration 2 ─────────────────────────────────────────────────────────────────
#> ℹ Using 10 of 16 coalitions, 2 new.
#>
#> ── Iteration 3 ─────────────────────────────────────────────────────────────────
#> ℹ Using 12 of 16 coalitions, 2 new.
# When not using all coalitions, we can also get the SD of the Shapley values,
# reflecting uncertainty in the coalition sampling part of the procedure.
print(explain_iterative, what = "shapley_sd")
#> explain_id none Solar.R Wind Temp Month
#> <int> <num> <num> <num> <num> <num>
#> 1: 1 0 0.381 1.63 1.89 0.771
#> 2: 2 0 0.428 2.69 2.64 0.907
#> 3: 3 0 0.423 3.09 3.01 0.978
## Summary
# For iterative estimation, convergence info is also provided
summary_iterative <- summary(explain_iterative)
# }